+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Feline Calicivirus Strain F9 | |||||||||
 Map data Map data | Icosahedral reconstruction of feline calicivirus strain F9 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Capsid / Calicivirus / Vesivirus / Vp1 / VIRUS | |||||||||
| Function / homology | T=3 icosahedral viral capsid / Calicivirus coat protein / Calicivirus coat protein / Picornavirus/Calicivirus coat protein / Viral coat protein subunit / host cell cytoplasm / Capsid protein VP1 Function and homology information Function and homology information | |||||||||
| Biological species |  Feline calicivirus Feline calicivirus | |||||||||
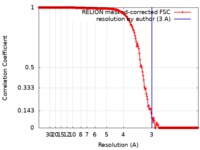
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Conley MJ / Bhella D | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Calicivirus VP2 forms a portal-like assembly following receptor engagement. Authors: Michaela J Conley / Marion McElwee / Liyana Azmi / Mads Gabrielsen / Olwyn Byron / Ian G Goodfellow / David Bhella /  Abstract: To initiate infection, many viruses enter their host cells by triggering endocytosis following receptor engagement. However, the mechanisms by which non-enveloped viruses escape the endosome are ...To initiate infection, many viruses enter their host cells by triggering endocytosis following receptor engagement. However, the mechanisms by which non-enveloped viruses escape the endosome are poorly understood. Here we present near-atomic-resolution cryo-electron microscopy structures for feline calicivirus both undecorated and labelled with a soluble fragment of its cellular receptor, feline junctional adhesion molecule A. We show that VP2, a minor capsid protein encoded by all caliciviruses, forms a large portal-like assembly at a unique three-fold axis of symmetry, following receptor engagement. This assembly-which was not detected in undecorated virions-is formed of twelve copies of VP2, arranged with their hydrophobic N termini pointing away from the virion surface. Local rearrangement at the portal site leads to the opening of a pore in the capsid shell. We hypothesize that the portal-like assembly functions as a channel for the delivery of the calicivirus genome, through the endosomal membrane, into the cytoplasm of a host cell, thereby initiating infection. VP2 was previously known to be critical for the production of infectious virus; our findings provide insights into its structure and function that advance our understanding of the Caliciviridae. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0054.map.gz emd_0054.map.gz | 480.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0054-v30.xml emd-0054-v30.xml emd-0054.xml emd-0054.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0054_fsc.xml emd_0054_fsc.xml | 17.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_0054.png emd_0054.png | 226.5 KB | ||
| Filedesc metadata |  emd-0054.cif.gz emd-0054.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0054 http://ftp.pdbj.org/pub/emdb/structures/EMD-0054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0054 | HTTPS FTP |
-Related structure data
| Related structure data |  6gshMC  0056C  6gsiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10192 (Title: Calicivirus VP2 forms a portal to mediate endosome escape EMPIAR-10192 (Title: Calicivirus VP2 forms a portal to mediate endosome escapeData size: 324.9 Data #1: Motion corrected micrographs of feline calicivirus strain F9 [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0054.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0054.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedral reconstruction of feline calicivirus strain F9 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : T=3 Icosahedral Capsid.
| Entire | Name: T=3 Icosahedral Capsid. |
|---|---|
| Components |
|
-Supramolecule #1: T=3 Icosahedral Capsid.
| Supramolecule | Name: T=3 Icosahedral Capsid. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: T=3 Icosahedral Capsid |
|---|---|
| Source (natural) | Organism:  Feline calicivirus Feline calicivirus |
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Feline calicivirus Feline calicivirus |
| Molecular weight | Theoretical: 73.346664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MCSTCANVLK YYNWDPHFRL VIDPNKFLSV GFCDNPLMCC YPELLPEFGT VWDCDQSPLQ IYLESILGDD EWASTYEAID PAVPPMHWD AAGKIFQPHP GVLMHHLIGE VAKAWDPNLP MFRLEADGDG SITAPEQGTP VGGVIAEPSA QMSAAADMAT G KSVDSEWE ...String: MCSTCANVLK YYNWDPHFRL VIDPNKFLSV GFCDNPLMCC YPELLPEFGT VWDCDQSPLQ IYLESILGDD EWASTYEAID PAVPPMHWD AAGKIFQPHP GVLMHHLIGE VAKAWDPNLP MFRLEADGDG SITAPEQGTP VGGVIAEPSA QMSAAADMAT G KSVDSEWE AFFSFHTSVN WSTSETQGKI LFKQSLGPLL NPYLEHLAKL YVAWSGSIDV RFSISGSGVF GGKLAAIVVP PG VDPVQST SMLQYPHVLF DARQVEPVIF SIPDLRSTLY HLMSDTDTTS LVIMVYNDLI NPYANDSNSS GCIVTVETKP GAD FKFHLL KPPGSMLTHG SVPSDLIPKS SSLWIGNRHW TDITDFVIRP FVFQANRHFD FNQETAGWST PRYRPITITI SEKN GAKLG IGVATDYIVP GIPDGWPDTT IPEKLTPAGD YAITNKSGND ITTAAGYDGA DVIVNNTNFK GMYICGSLQR AWGDK KISN TAFITTATKV DNAIEPSNVI DMTKIAVYQD THVGKEVQTS DDTLSLLGYT GIGEQAIGSD RDRVVRISVL PETGAR GGN HPIFYKNSIK LGYVIRSIDV FNSQILHTSR QLSLNHYLLP PDSFAVYRII DSNGSWFDIG IDSDGFSFVG VSSIGKL EF PLTASYMGIQ LAKIRLASNI RSSMTKL UniProtKB: Capsid protein VP1 |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 3 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.2 / Details: Phosphate buffered saline |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | Purified enveloped virions |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 5198 / Average electron dose: 63.0 e/Å2 Details: Each micrograph was recorded as a movie of 50 individual fractions with a total dose of 63 e/angstrom squared |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 75000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)