[English] 日本語
 Yorodumi
Yorodumi- EMDB-0030: Structure of activated transcription complex Pol II-DSIF-PAF-SPT6... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of activated transcription complex Pol II-DSIF-PAF-SPT6 (Map B, NGN/SPT4 selected particles) | |||||||||
 Map data Map data | Global refinement of NGN/SPT4 selected particles. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
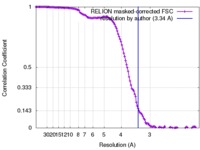
| Method | single particle reconstruction / cryo EM / Resolution: 3.34 Å | |||||||||
 Authors Authors | Vos SM / Farnung L / Boehning M / Linden A / Wigge C / Urlaub H / Cramer P | |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Authors: Seychelle M Vos / Lucas Farnung / Marc Boehning / Christoph Wigge / Andreas Linden / Henning Urlaub / Patrick Cramer /  Abstract: Gene regulation involves activation of RNA polymerase II (Pol II) that is paused and bound by the protein complexes DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF). Here ...Gene regulation involves activation of RNA polymerase II (Pol II) that is paused and bound by the protein complexes DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF). Here we show that formation of an activated Pol II elongation complex in vitro requires the kinase function of the positive transcription elongation factor b (P-TEFb) and the elongation factors PAF1 complex (PAF) and SPT6. The cryo-EM structure of an activated elongation complex of Sus scrofa Pol II and Homo sapiens DSIF, PAF and SPT6 was determined at 3.1 Å resolution and compared to the structure of the paused elongation complex formed by Pol II, DSIF and NELF. PAF displaces NELF from the Pol II funnel for pause release. P-TEFb phosphorylates the Pol II linker to the C-terminal domain. SPT6 binds to the phosphorylated C-terminal-domain linker and opens the RNA clamp formed by DSIF. These results provide the molecular basis for Pol II pause release and elongation activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0030.map.gz emd_0030.map.gz | 140.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0030-v30.xml emd-0030-v30.xml emd-0030.xml emd-0030.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0030_fsc.xml emd_0030_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0030.png emd_0030.png | 70.6 KB | ||
| Masks |  emd_0030_msk_1.map emd_0030_msk_1.map | 178 MB |  Mask map Mask map | |
| Others |  emd_0030_additional.map.gz emd_0030_additional.map.gz emd_0030_half_map_1.map.gz emd_0030_half_map_1.map.gz emd_0030_half_map_2.map.gz emd_0030_half_map_2.map.gz | 16.7 MB 140.7 MB 140.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0030 http://ftp.pdbj.org/pub/emdb/structures/EMD-0030 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0030 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0030 | HTTPS FTP |
-Related structure data
| Related structure data |  0031C  0032C  0033C  0034C  0035C  0036C  0037C  6gmeC  6gmhC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0030.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0030.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Global refinement of NGN/SPT4 selected particles. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.049 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_0030_msk_1.map emd_0030_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Postprocessed map of global refinement for NGN/SPT4 selected...
| File | emd_0030_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map of global refinement for NGN/SPT4 selected particles with an applied B factor of -90.81 (Map B). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 of global refinement for NGN/SPT4...
| File | emd_0030_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of global refinement for NGN/SPT4 selected particles (Map B). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 of global refinement for NGN/SPT4...
| File | emd_0030_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of global refinement for NGN/SPT4 selected particles (Map B). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RNA Polymerase II-DSIF-PAF-SPT6 elongation complex (EC*)
| Entire | Name: RNA Polymerase II-DSIF-PAF-SPT6 elongation complex (EC*) |
|---|---|
| Components |
|
-Supramolecule #1: RNA Polymerase II-DSIF-PAF-SPT6 elongation complex (EC*)
| Supramolecule | Name: RNA Polymerase II-DSIF-PAF-SPT6 elongation complex (EC*) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#21, #23 |
|---|---|
| Molecular weight | Theoretical: 1.257 MDa |
-Supramolecule #2: RNA Polymerase II
| Supramolecule | Name: RNA Polymerase II / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#9, #11, #10, #12 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: associated proteins
| Supramolecule | Name: associated proteins / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #13, #16, #18, #23, #19-#22 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Supramolecule #4: Nucleic acids
| Supramolecule | Name: Nucleic acids / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #14-#15, #17 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 10s wait prior to blotting, blotting 8.5s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 10.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)