[English] 日本語
 Yorodumi
Yorodumi- EMDB-0013: Helical reconstruction of BCL10 CARD and MALT1 DEATH DOMAIN complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical reconstruction of BCL10 CARD and MALT1 DEATH DOMAIN complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | BCL10 / MALT1 / CBM complex / helical reconstruction / cancer / autoimmune disease / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of lymphotoxin A production / polkadots / B-1 B cell differentiation / positive regulation of T-helper 17 cell differentiation / CBM complex / regulation of T cell receptor signaling pathway / protein kinase B binding / antifungal innate immune response / response to fungus / positive regulation of mast cell cytokine production ...positive regulation of lymphotoxin A production / polkadots / B-1 B cell differentiation / positive regulation of T-helper 17 cell differentiation / CBM complex / regulation of T cell receptor signaling pathway / protein kinase B binding / antifungal innate immune response / response to fungus / positive regulation of mast cell cytokine production / CARD domain binding / T cell apoptotic process / negative regulation of mature B cell apoptotic process / B cell apoptotic process / CLEC7A/inflammasome pathway / programmed cell death / nuclear export / positive regulation of extrinsic apoptotic signaling pathway / toll-like receptor signaling pathway / response to food / non-canonical NF-kappaB signal transduction / small molecule binding / positive regulation of T cell receptor signaling pathway / immunoglobulin mediated immune response / B cell activation / general transcription initiation factor binding / immunological synapse / positive regulation of phosphorylation / NF-kappaB binding / cellular defense response / endopeptidase activator activity / T cell proliferation / cytoplasmic microtubule / signaling adaptor activity / positive regulation of interleukin-2 production / : / lipopolysaccharide-mediated signaling pathway / positive regulation of protein ubiquitination / positive regulation of interleukin-1 beta production / positive regulation of interleukin-8 production / neural tube closure / apoptotic signaling pathway / cellular response to mechanical stimulus / Activation of NF-kappaB in B cells / defense response / : / positive regulation of T cell cytokine production / protein homooligomerization / positive regulation of interleukin-6 production / CLEC7A (Dectin-1) signaling / FCERI mediated NF-kB activation / fibrillar center / positive regulation of T cell activation / ubiquitin-protein transferase activity / peptidase activity / Downstream TCR signaling / T cell receptor signaling pathway / E3 ubiquitin ligases ubiquitinate target proteins / cellular response to lipopolysaccharide / protease binding / endopeptidase activity / regulation of apoptotic process / protein-macromolecule adaptor activity / adaptive immune response / transcription coactivator activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / positive regulation of canonical NF-kappaB signal transduction / lysosome / positive regulation of apoptotic process / membrane raft / innate immune response / cysteine-type endopeptidase activity / ubiquitin protein ligase binding / negative regulation of apoptotic process / positive regulation of DNA-templated transcription / protein-containing complex binding / perinuclear region of cytoplasm / protein-containing complex / proteolysis / nucleoplasm / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
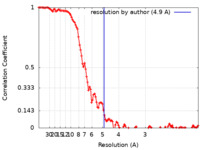
| Method | helical reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Schlauderer F / Desfosses A | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Molecular architecture and regulation of BCL10-MALT1 filaments. Authors: Florian Schlauderer / Thomas Seeholzer / Ambroise Desfosses / Torben Gehring / Mike Strauss / Karl-Peter Hopfner / Irina Gutsche / Daniel Krappmann / Katja Lammens /    Abstract: The CARD11-BCL10-MALT1 (CBM) complex triggers the adaptive immune response in lymphocytes and lymphoma cells. CARD11/CARMA1 acts as a molecular seed inducing BCL10 filaments, but the integration of ...The CARD11-BCL10-MALT1 (CBM) complex triggers the adaptive immune response in lymphocytes and lymphoma cells. CARD11/CARMA1 acts as a molecular seed inducing BCL10 filaments, but the integration of MALT1 and the assembly of a functional CBM complex has remained elusive. Using cryo-EM we solved the helical structure of the BCL10-MALT1 filament. The structural model of the filament core solved at 4.9 Å resolution identified the interface between the N-terminal MALT1 DD and the BCL10 caspase recruitment domain. The C-terminal MALT1 Ig and paracaspase domains protrude from this core to orchestrate binding of mediators and substrates at the filament periphery. Mutagenesis studies support the importance of the identified BCL10-MALT1 interface for CBM complex assembly, MALT1 protease activation and NF-κB signaling in Jurkat and primary CD4 T-cells. Collectively, we present a model for the assembly and architecture of the CBM signaling complex and how it functions as a signaling hub in T-lymphocytes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0013.map.gz emd_0013.map.gz | 66.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0013-v30.xml emd-0013-v30.xml emd-0013.xml emd-0013.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0013_fsc.xml emd_0013_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_0013.png emd_0013.png | 274.1 KB | ||
| Filedesc metadata |  emd-0013.cif.gz emd-0013.cif.gz | 5.3 KB | ||
| Others |  emd_0013_half_map_1.map.gz emd_0013_half_map_1.map.gz emd_0013_half_map_2.map.gz emd_0013_half_map_2.map.gz | 46 MB 46 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0013 http://ftp.pdbj.org/pub/emdb/structures/EMD-0013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0013 | HTTPS FTP |
-Related structure data
| Related structure data |  6gk2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0013.map.gz / Format: CCP4 / Size: 91.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0013.map.gz / Format: CCP4 / Size: 91.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.002 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Unmasked, unfiltered, helically symmetrized half map 2
| File | emd_0013_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked, unfiltered, helically symmetrized half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unmasked, unfiltered, helically symmetrized half map 1
| File | emd_0013_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked, unfiltered, helically symmetrized half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of BCL10 CARD and MALT1 DEATH DOMAIN
| Entire | Name: Complex of BCL10 CARD and MALT1 DEATH DOMAIN |
|---|---|
| Components |
|
-Supramolecule #1: Complex of BCL10 CARD and MALT1 DEATH DOMAIN
| Supramolecule | Name: Complex of BCL10 CARD and MALT1 DEATH DOMAIN / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 104 kDa/nm |
-Macromolecule #1: B-cell lymphoma/leukemia 10
| Macromolecule | Name: B-cell lymphoma/leukemia 10 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.614566 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EEDLTEVKKD ALENLRVYLC EKIIAERHFD HLRAKKILSR EDTEEISCRT SSRKRAGKLL DYLQENPKGL DTLVESIRRE KTQNFLIQK ITDEVLKLRN IKLEHLK UniProtKB: B-cell lymphoma/leukemia 10 |
-Macromolecule #2: Mucosa-associated lymphoid tissue lymphoma translocation protein 1
| Macromolecule | Name: Mucosa-associated lymphoid tissue lymphoma translocation protein 1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.401128 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LNRLREPLLR RLSELLDQAP EGRGWRRLAE LAGSRGRLRL SCLDLEQCSL KVLEPEGSPS LCLLKLMGEK GCTVTELSDF LQAMEHTEV LQL UniProtKB: Mucosa-associated lymphoid tissue lymphoma translocation protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 99.6 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)