[English] 日本語
 Yorodumi
Yorodumi- SASDGE4: Mixed lineage leukemia protein-1 complex, MLL1-WDR5-ASH2L-RBBP5(2-480) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | Mixed lineage leukemia protein-1 complex, MLL1-WDR5-ASH2L-RBBP5(2-480)
|
| Function / homology |  Function and homology information Function and homology informationnegative regulation of DNA methylation-dependent heterochromatin formation / protein-cysteine methyltransferase activity / [histone H3]-lysine4 N-methyltransferase / histone H3K4 monomethyltransferase activity / response to potassium ion / unmethylated CpG binding / histone H3K4 trimethyltransferase activity / histone H3Q5ser reader activity / histone H3K4me1 reader activity / Epigenetic regulation of gene expression by MLL3 and MLL4 complexes ...negative regulation of DNA methylation-dependent heterochromatin formation / protein-cysteine methyltransferase activity / [histone H3]-lysine4 N-methyltransferase / histone H3K4 monomethyltransferase activity / response to potassium ion / unmethylated CpG binding / histone H3K4 trimethyltransferase activity / histone H3Q5ser reader activity / histone H3K4me1 reader activity / Epigenetic regulation of gene expression by MLL3 and MLL4 complexes / MLL3/4 complex / Set1C/COMPASS complex / MLL1/2 complex / definitive hemopoiesis / ATAC complex / regulation of short-term neuronal synaptic plasticity / NSL complex / histone H3K4 methyltransferase activity / Cardiogenesis / anterior/posterior pattern specification / T-helper 2 cell differentiation / embryonic hemopoiesis / Formation of WDR5-containing histone-modifying complexes / exploration behavior / minor groove of adenine-thymine-rich DNA binding / histone methyltransferase complex / MLL1 complex / regulation of cell division / regulation of embryonic development / histone acetyltransferase complex / membrane depolarization / cellular response to transforming growth factor beta stimulus / negative regulation of fibroblast proliferation / spleen development / positive regulation of gluconeogenesis / transcription initiation-coupled chromatin remodeling / homeostasis of number of cells within a tissue / Transferases; Transferring one-carbon groups; Methyltransferases / post-embryonic development / gluconeogenesis / skeletal system development / circadian regulation of gene expression / Deactivation of the beta-catenin transactivating complex / Formation of the beta-catenin:TCF transactivating complex / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / visual learning / protein modification process / PKMTs methylate histone lysines / response to estrogen / Activation of anterior HOX genes in hindbrain development during early embryogenesis / RMTs methylate histone arginines / Transcriptional regulation of granulopoiesis / mitotic spindle / HATs acetylate histones / RUNX1 regulates transcription of genes involved in differentiation of HSCs / Neddylation / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / protein-containing complex assembly / fibroblast proliferation / histone binding / methylation / transcription cis-regulatory region binding / regulation of cell cycle / apoptotic process / DNA damage response / chromatin binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / nucleolus / negative regulation of transcription by RNA polymerase II / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / zinc ion binding / nucleoplasm / identical protein binding / nucleus / cytosol Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2019 Journal: Nucleic Acids Res / Year: 2019Title: The internal interaction in RBBP5 regulates assembly and activity of MLL1 methyltransferase complex. Authors: Jianming Han / Tingting Li / Yanjing Li / Muchun Li / Xiaoman Wang / Chao Peng / Chen Su / Na Li / Yiwen Li / Ying Xu / Yong Chen /  Abstract: The Mixed Lineage Leukemia protein 1 (MLL1) plays an essential role in the maintenance of the histone H3 lysine 4 (H3K4) methylation status for gene expression during differentiation and development. ...The Mixed Lineage Leukemia protein 1 (MLL1) plays an essential role in the maintenance of the histone H3 lysine 4 (H3K4) methylation status for gene expression during differentiation and development. The methyltransferase activity of MLL1 is regulated by three conserved core subunits, WDR5, RBBP5 and ASH2L. Here, we determined the structure of human RBBP5 and demonstrated its role in the assembly and regulation of the MLL1 complex. We identified an internal interaction between the WD40 propeller and the C-terminal distal region in RBBP5, which assisted the maintenance of the compact conformation of the MLL1 complex. We also discovered a vertebrate-specific motif in the C-terminal distal region of RBBP5 that contributed to nucleosome recognition and methylation of nucleosomes by the MLL1 complex. Our results provide new insights into functional conservation and evolutionary plasticity of the scaffold protein RBBP5 in the regulation of KMT2-family methyltransferase complexes. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDGE4 SASDGE4 |
|---|
-Related structure data
| Related structure data |  6km7C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Mixed lineage leukemia protein-1 complex, MLL1-WDR5-ASH2L-RBBP5(2-480) Specimen concentration: 2 mg/ml / Entity id: 1862 / 1863 / 1864 / 1865 |
|---|---|
| Buffer | Name: 300 mM NaCl, 25mM Tris-HCl, 4% glycerol, 1 mM TCEP / pH: 8 / Comment: degassing treatment |
| Entity #1862 | Name: MLL1 / Type: protein / Description: Histone-lysine N-methyltransferase 2A / Formula weight: 24.927 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q03164 Sequence: NEPPLNPHGS ARAEVHLRKS AFDMFNFLAS KHRQPPEYNP NDEEEEEVQL KSARRATSMD LPMPMRFRHL KKTSKEAVGV YRSPIHGRGL FCKRNIDAGE MVIEYAGNVI RSIQTDKREK YYDSKGIGCY MFRIDDSEVV DATMHGNAAR FINHSCEPNC YSRVINIDGQ ...Sequence: NEPPLNPHGS ARAEVHLRKS AFDMFNFLAS KHRQPPEYNP NDEEEEEVQL KSARRATSMD LPMPMRFRHL KKTSKEAVGV YRSPIHGRGL FCKRNIDAGE MVIEYAGNVI RSIQTDKREK YYDSKGIGCY MFRIDDSEVV DATMHGNAAR FINHSCEPNC YSRVINIDGQ KHIVIFAMRK IYRGEELTYD YKFPIEDASN KLPCNCGAKK CRKFLN |
| Entity #1863 | Name: WDR5 / Type: protein / Description: WD repeat-containing protein 5 / Formula weight: 34.26 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: P61964 Sequence: ATQSKPTPVK PNYALKFTLA GHTKAVSSVK FSPNGEWLAS SSADKLIKIW GAYDGKFEKT ISGHKLGISD VAWSSDSNLL VSASDDKTLK IWDVSSGKCL KTLKGHSNYV FCCNFNPQSN LIVSGSFDES VRIWDVKTGK CLKTLPAHSD PVSAVHFNRD GSLIVSSSYD ...Sequence: ATQSKPTPVK PNYALKFTLA GHTKAVSSVK FSPNGEWLAS SSADKLIKIW GAYDGKFEKT ISGHKLGISD VAWSSDSNLL VSASDDKTLK IWDVSSGKCL KTLKGHSNYV FCCNFNPQSN LIVSGSFDES VRIWDVKTGK CLKTLPAHSD PVSAVHFNRD GSLIVSSSYD GLCRIWDTAS GQCLKTLIDD DNPPVSFVKF SPNGKYILAA TLDNTLKLWD YSKGKCLKTY TGHKNEKYCI FANFSVTGGK WIVSGSEDNL VYIWNLQTKE IVQKLQGHTD VVISTACHPT ENIIASAALE NDKTIKLWKS DC |
| Entity #1864 | Name: ASH2L / Type: protein Description: Set1/Ash2 histone methyltransferase complex subunit ASH2 Formula weight: 60.207 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q9UBL3-3 Sequence: MDTQAGSVDE ENGRQLGEVE LQCGICTKWF TADTFGIDTS SCLPFMTNYS FHCNVCHHSG NTYFLRKQAN LKEMCLSALA NLTWQSRTQD EHPKTMFSKD KDIIPFIDKY WECMTTRQRP GKMTWPNNIV KTMSKERDVF LVKEHPDPGS KDPEEDYPKF GLLDQDLSNI ...Sequence: MDTQAGSVDE ENGRQLGEVE LQCGICTKWF TADTFGIDTS SCLPFMTNYS FHCNVCHHSG NTYFLRKQAN LKEMCLSALA NLTWQSRTQD EHPKTMFSKD KDIIPFIDKY WECMTTRQRP GKMTWPNNIV KTMSKERDVF LVKEHPDPGS KDPEEDYPKF GLLDQDLSNI GPAYDNQKQS SAVSTSGNLN GGIAAGSSGK GRGAKRKQQD GGTTGTTKKA RSDPLFSAQR LPPHGYPLEH PFNKDGYRYI LAEPDPHAPD PEKLELDCWA GKPIPGDLYR ACLYERVLLA LHDRAPQLKI SDDRLTVVGE KGYSMVRASH GVRKGAWYFE ITVDEMPPDT AARLGWSQPL GNLQAPLGYD KFSYSWRSKK GTKFHQSIGK HYSSGYGQGD VLGFYINLPE DTETAKSLPD TYKDKALIKF KSYLYFEEKD FVDKAEKSLK QTPHSEIIFY KNGVNQGVAY KDIFEGVYFP AISLYKSCTV SINFGPCFKY PPKDLTYRPM SDMGWGAVVE HTLADVLYHV ETEVDGRRSP PWEP |
| Entity #1865 | Name: RBBP5 / Type: protein / Description: Retinoblastoma-binding protein 5 / Formula weight: 52.962 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q15291 Sequence: NLELLESFGQ NYPEEADGTL DCISMALTCT FNRWGTLLAV GCNDGRIVIW DFLTRGIAKI ISAHIHPVCS LCWSRDGHKL VSASTDNIVS QWDVLSGDCD QRFRFPSPIL KVQYHPRDQN KVLVCPMKSA PVMLTLSDSK HVVLPVDDDS DLNVVASFDR RGEYIYTGNA ...Sequence: NLELLESFGQ NYPEEADGTL DCISMALTCT FNRWGTLLAV GCNDGRIVIW DFLTRGIAKI ISAHIHPVCS LCWSRDGHKL VSASTDNIVS QWDVLSGDCD QRFRFPSPIL KVQYHPRDQN KVLVCPMKSA PVMLTLSDSK HVVLPVDDDS DLNVVASFDR RGEYIYTGNA KGKILVLKTD SQDLVASFRV TTGTSNTTAI KSIEFARKGS CFLINTADRI IRVYDGREIL TCGRDGEPEP MQKLQDLVNR TPWKKCCFSG DGEYIVAGSA RQHALYIWEK SIGNLVKILH GTRGELLLDV AWHPVRPIIA SISSGVVSIW AQNQVENWSA FAPDFKELDE NVEYEERESE FDIEDEDKSE PEQTGADAAE DEEVDVTSVD PIAAFCSSDE ELEDSKALLY LPIAPEVEDP EENPYGPPPD AVQTSLMDEG ASSEKKRQSS ADGSQPPKKK PKTTNIELQG VPNDEVHPLL GVKGDGKSK |
-Experimental information
| Beam | Instrument name: Shanghai Synchrotron Radiation Facility (SSRF) BL19U2 City: Shanghai / 国: China  / Type of source: X-ray synchrotron / Wavelength: 0.0918 Å / Dist. spec. to detc.: 2.415 mm / Type of source: X-ray synchrotron / Wavelength: 0.0918 Å / Dist. spec. to detc.: 2.415 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | ||||||||||||||||||||||||||||||
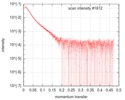
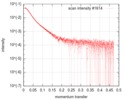
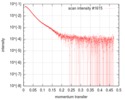
| Scan | Measurement date: Jun 22, 2019 / Storage temperature: 10 °C / Cell temperature: 10 °C / Exposure time: 1 sec. / Number of frames: 20 / Unit: 1/A /
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller