[English] 日本語
 Yorodumi
Yorodumi- SASDFE2: wild-type human Latency Associated Peptide (LAP) (Latency Associa... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 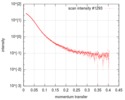 |
|---|---|
 Sample Sample | wild-type human Latency Associated Peptide (LAP)
|
| Function / homology |  Function and homology information Function and homology informationcellular response to acetaldehyde / frontal suture morphogenesis / Influenza Virus Induced Apoptosis / adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains / positive regulation of microglia differentiation / regulation of interleukin-23 production / branch elongation involved in mammary gland duct branching / positive regulation of primary miRNA processing / columnar/cuboidal epithelial cell maturation / negative regulation of skeletal muscle tissue development ...cellular response to acetaldehyde / frontal suture morphogenesis / Influenza Virus Induced Apoptosis / adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains / positive regulation of microglia differentiation / regulation of interleukin-23 production / branch elongation involved in mammary gland duct branching / positive regulation of primary miRNA processing / columnar/cuboidal epithelial cell maturation / negative regulation of skeletal muscle tissue development / macrophage derived foam cell differentiation / response to laminar fluid shear stress / embryonic liver development / regulation of enamel mineralization / regulation of branching involved in mammary gland duct morphogenesis / regulation of cartilage development / TGFBR2 MSI Frameshift Mutants in Cancer / regulation of striated muscle tissue development / regulation of blood vessel remodeling / regulation of protein import into nucleus / regulatory T cell differentiation / tolerance induction to self antigen / extracellular matrix assembly / negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / negative regulation of hyaluronan biosynthetic process / type III transforming growth factor beta receptor binding / myofibroblast differentiation / positive regulation of cardiac muscle cell differentiation / positive regulation of odontogenesis / connective tissue replacement involved in inflammatory response wound healing / Langerhans cell differentiation / TGFBR2 Kinase Domain Mutants in Cancer / positive regulation of smooth muscle cell differentiation / positive regulation of exit from mitosis / negative regulation of macrophage cytokine production / odontoblast differentiation / SMAD2/3 Phosphorylation Motif Mutants in Cancer / TGFBR1 KD Mutants in Cancer / positive regulation of mesenchymal stem cell proliferation / positive regulation of isotype switching to IgA isotypes / positive regulation of receptor signaling pathway via STAT / membrane protein intracellular domain proteolysis / positive regulation of extracellular matrix assembly / retina vasculature development in camera-type eye / heart valve morphogenesis / bronchiole development / hyaluronan catabolic process / mammary gland branching involved in thelarche / TGFBR3 regulates TGF-beta signaling / positive regulation of vasculature development / lens fiber cell differentiation / ATP biosynthetic process / type II transforming growth factor beta receptor binding / negative regulation of extracellular matrix disassembly / positive regulation of branching involved in ureteric bud morphogenesis / receptor catabolic process / TGFBR1 LBD Mutants in Cancer / type I transforming growth factor beta receptor binding / positive regulation of chemotaxis / response to salt / germ cell migration / positive regulation of mononuclear cell migration / endoderm development / phospholipid homeostasis / negative regulation of cell-cell adhesion mediated by cadherin / negative regulation of myoblast differentiation / positive regulation of vascular permeability / negative regulation of biomineral tissue development / response to cholesterol / oligodendrocyte development / negative regulation of interleukin-17 production / cell-cell junction organization / phosphate-containing compound metabolic process / surfactant homeostasis / deubiquitinase activator activity / sprouting angiogenesis / negative regulation of release of sequestered calcium ion into cytosol / positive regulation of chemokine (C-X-C motif) ligand 2 production / negative regulation of ossification / digestive tract development / aortic valve morphogenesis / RUNX3 regulates CDKN1A transcription / response to vitamin D / positive regulation of fibroblast migration / face morphogenesis / ureteric bud development / positive regulation of epidermal growth factor receptor signaling pathway / neural tube development / positive regulation of regulatory T cell differentiation / positive regulation of peptidyl-tyrosine phosphorylation / Molecules associated with elastic fibres / negative regulation of neuroblast proliferation / lung alveolus development / negative regulation of phagocytosis / Syndecan interactions / ventricular cardiac muscle tissue morphogenesis / muscle cell cellular homeostasis / cellular response to insulin-like growth factor stimulus / negative regulation of fat cell differentiation / odontogenesis of dentin-containing tooth Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Date: 2019 Jul 1 Date: 2019 Jul 1Title: Structural consequences of transforming growth factor beta-1 activation from near-therapeutic X-ray doses Authors: Stachowski T / Grant T |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDFE2 SASDFE2 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: wild-type human Latency Associated Peptide (LAP) / Specimen concentration: 4.5 mg/ml |
|---|---|
| Buffer | Name: phosphate buffered saline / pH: 7.4 |
| Entity #1479 | Name: LAP / Type: protein / Description: Latency Associated Peptide / Formula weight: 29.212 / Num. of mol.: 2 / Source: Homo sapiens / References: UniProt: P01137 Sequence: LSTSKTIDME LVKRKRIEAI RGQILSKLRL ASPPSQGEVP PGPLPEAVLA LYNSTRDRVA GESAEPEPEP EADYYAKEVT RVLMVETHNE IYDKFKQSTH SIYMFFNTSE LREAVPEPVL LSRAELRLLR LKLKVEQHVE LYQKYSNNSW RYLSNRLLAP SDSPEWLSFD ...Sequence: LSTSKTIDME LVKRKRIEAI RGQILSKLRL ASPPSQGEVP PGPLPEAVLA LYNSTRDRVA GESAEPEPEP EADYYAKEVT RVLMVETHNE IYDKFKQSTH SIYMFFNTSE LREAVPEPVL LSRAELRLLR LKLKVEQHVE LYQKYSNNSW RYLSNRLLAP SDSPEWLSFD VTGVVRQWLS RGGEIEGFRL SAHCSCDSRD NTLQVDINGF TTGRRGDLAT IHGMNRPFLL LMATPLERAQ HLQSSRHRAH HHHHH |
-Experimental information
| Beam | Instrument name: Advanced Light Source (ALS) 12.3.1 (SIBYLS) City: Berkeley, CA / 国: USA  / Type of source: X-ray synchrotron / Wavelength: 0.1127 Å / Dist. spec. to detc.: 2 mm / Type of source: X-ray synchrotron / Wavelength: 0.1127 Å / Dist. spec. to detc.: 2 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus3 X 2M / Pixsize x: 172 mm | ||||||||||||||||||||||||||||||
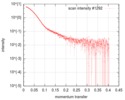
| Scan |
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller