+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  |
|---|---|
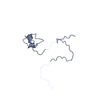
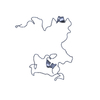
 試料 試料 | Labeled Importin Beta Binding Domain from importin subunit alpha-1 (IBB-Alexa488/Alexa594) with denaturant
|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Sensing of DNA Double Strand Breaks / regulation of DNA recombination / entry of viral genome into host nucleus through nuclear pore complex via importin / positive regulation of viral life cycle / NLS-dependent protein nuclear import complex / NS1 Mediated Effects on Host Pathways / NLS-bearing protein import into nucleus / nuclear localization sequence binding / nuclear import signal receptor activity / CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde ...Sensing of DNA Double Strand Breaks / regulation of DNA recombination / entry of viral genome into host nucleus through nuclear pore complex via importin / positive regulation of viral life cycle / NLS-dependent protein nuclear import complex / NS1 Mediated Effects on Host Pathways / NLS-bearing protein import into nucleus / nuclear localization sequence binding / nuclear import signal receptor activity / CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde / CaMK IV-mediated phosphorylation of CREB / DNA metabolic process / positive regulation of type I interferon production / ISG15 antiviral mechanism / SARS-CoV-1 activates/modulates innate immune responses / histone deacetylase binding / protein import into nucleus / host cell / nuclear membrane / Estrogen-dependent gene expression / Golgi membrane / endoplasmic reticulum membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / RNA binding / nucleoplasm / nucleus / membrane / cytosol / cytoplasm 類似検索 - 分子機能 |
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) |
 引用 引用 |  ジャーナル: Proc Natl Acad Sci U S A / 年: 2017 ジャーナル: Proc Natl Acad Sci U S A / 年: 2017タイトル: Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. 著者: Gustavo Fuertes / Niccolò Banterle / Kiersten M Ruff / Aritra Chowdhury / Davide Mercadante / Christine Koehler / Michael Kachala / Gemma Estrada Girona / Sigrid Milles / Ankur Mishra / ...著者: Gustavo Fuertes / Niccolò Banterle / Kiersten M Ruff / Aritra Chowdhury / Davide Mercadante / Christine Koehler / Michael Kachala / Gemma Estrada Girona / Sigrid Milles / Ankur Mishra / Patrick R Onck / Frauke Gräter / Santiago Esteban-Martín / Rohit V Pappu / Dmitri I Svergun / Edward A Lemke /     要旨: Unfolded states of proteins and native states of intrinsically disordered proteins (IDPs) populate heterogeneous conformational ensembles in solution. The average sizes of these heterogeneous ...Unfolded states of proteins and native states of intrinsically disordered proteins (IDPs) populate heterogeneous conformational ensembles in solution. The average sizes of these heterogeneous systems, quantified by the radius of gyration ( ), can be measured by small-angle X-ray scattering (SAXS). Another parameter, the mean dye-to-dye distance ( ) for proteins with fluorescently labeled termini, can be estimated using single-molecule Förster resonance energy transfer (smFRET). A number of studies have reported inconsistencies in inferences drawn from the two sets of measurements for the dimensions of unfolded proteins and IDPs in the absence of chemical denaturants. These differences are typically attributed to the influence of fluorescent labels used in smFRET and to the impact of high concentrations and averaging features of SAXS. By measuring the dimensions of a collection of labeled and unlabeled polypeptides using smFRET and SAXS, we directly assessed the contributions of dyes to the experimental values and For chemically denatured proteins we obtain mutual consistency in our inferences based on and , whereas for IDPs under native conditions, we find substantial deviations. Using computations, we show that discrepant inferences are neither due to methodological shortcomings of specific measurements nor due to artifacts of dyes. Instead, our analysis suggests that chemical heterogeneity in heteropolymeric systems leads to a decoupling between and that is amplified in the absence of denaturants. Therefore, joint assessments of and combined with measurements of polymer shapes should provide a consistent and complete picture of the underlying ensembles. |
 登録者 登録者 |
|
- 構造の表示
構造の表示
- ダウンロードとリンク
ダウンロードとリンク
-Data source
| SASBDBのページ |  SASDEU2 SASDEU2 |
|---|
-関連構造データ
| 関連構造データ | C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- 外部リンク
外部リンク
| 「今月の分子」の関連する項目 |
|---|
-モデル
- 試料
試料
 試料 試料 | 名称: Labeled Importin Beta Binding Domain from importin subunit alpha-1 (IBB-Alexa488/Alexa594) with denaturant 試料濃度: 10 mg/ml / Entity id: 1236 / 1252 / 1253 |
|---|---|
| バッファ | 名称: PBS, 10 mM DTT, 6 M urea, 0.3 M KCl / pH: 7.4 |
| 要素 #1236 | 名称: IBB-488/594 / タイプ: protein / 記述: Importin subunit alpha-1 / 分子量: 11.275 / 分子数: 1 / 由来: Homo sapiens / 参照: UniProt: P52292 配列: GCTNENANTP AARLHRFKNK GKDSTEMRRR RIEVNVELRK AKKDDQMLKR RNVSSFPDDA TSPLQENRNN QGTVNWSVDD IVKGINSSNV ENQLQATUA |
| 要素 #1252 | 名称: Alexa594 / タイプ: other / 記述: Alexa Fluor™ 594 C5 Maleimide / 分子量: 0.886 / 分子数: 1 |
| 要素 #1253 | 名称: Alexa488 / タイプ: other / 記述: Alexa Fluor™ 488 C5 Maleimide / 分子量: 0.7 / 分子数: 1 |
-実験情報
| ビーム | 設備名称: PETRA III EMBL P12 / 地域: Hamburg / 国: Germany  / 線源: X-ray synchrotron / 波長: 0.1 Å / スペクトロメータ・検出器間距離: 3 mm / 線源: X-ray synchrotron / 波長: 0.1 Å / スペクトロメータ・検出器間距離: 3 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 検出器 | 名称: Pilatus 2M | |||||||||||||||||||||||||||||||||
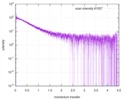
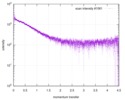
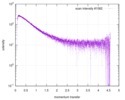
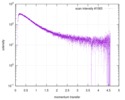
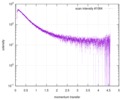
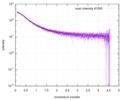
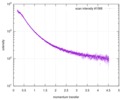
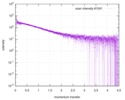
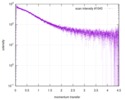
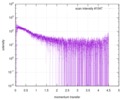
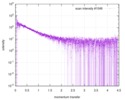
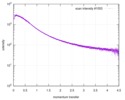
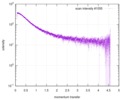
| スキャン | 測定日: 2013年6月15日 / セル温度: 23 °C / 照射時間: 0.05 sec. / フレーム数: 20 / 単位: 1/nm /
| |||||||||||||||||||||||||||||||||
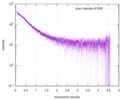
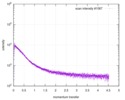
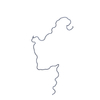
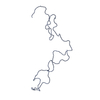
| 距離分布関数 P(R) |
| |||||||||||||||||||||||||||||||||
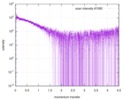
| 結果 | コメント: The protein contains a penultimate non-canonical amino acid p-acetylphenylalanine (207 Da) that is represented as U (selenocysteine, 168 Da) in the amino acid sequence for the entry. ...コメント: The protein contains a penultimate non-canonical amino acid p-acetylphenylalanine (207 Da) that is represented as U (selenocysteine, 168 Da) in the amino acid sequence for the entry. Therefore, the calculated MW from sequence (MW(expected)) must be adjusted accordingly (ca. 40 Da).
|
 ムービー
ムービー コントローラー
コントローラー