+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDEE2 |
|---|---|
 Sample Sample | The N-terminal domain of estrogen receptor alpha
|
| Function / homology |  Function and homology information Function and homology informationregulation of epithelial cell apoptotic process / antral ovarian follicle growth / regulation of branching involved in prostate gland morphogenesis / RUNX1 regulates transcription of genes involved in WNT signaling / RUNX1 regulates estrogen receptor mediated transcription / regulation of toll-like receptor signaling pathway / nuclear estrogen receptor activity / epithelial cell development / steroid hormone receptor signaling pathway / prostate epithelial cord elongation ...regulation of epithelial cell apoptotic process / antral ovarian follicle growth / regulation of branching involved in prostate gland morphogenesis / RUNX1 regulates transcription of genes involved in WNT signaling / RUNX1 regulates estrogen receptor mediated transcription / regulation of toll-like receptor signaling pathway / nuclear estrogen receptor activity / epithelial cell development / steroid hormone receptor signaling pathway / prostate epithelial cord elongation / epithelial cell proliferation involved in mammary gland duct elongation / prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis / mammary gland branching involved in pregnancy / uterus development / negative regulation of smooth muscle cell apoptotic process / vagina development / TFIIB-class transcription factor binding / androgen metabolic process / mammary gland alveolus development / cellular response to estrogen stimulus / estrogen response element binding / Mitochondrial unfolded protein response (UPRmt) / nuclear receptor-mediated steroid hormone signaling pathway / Nuclear signaling by ERBB4 / : / : / RNA polymerase II preinitiation complex assembly / positive regulation of nitric-oxide synthase activity / estrogen receptor signaling pathway / protein localization to chromatin / steroid binding / 14-3-3 protein binding / TFAP2 (AP-2) family regulates transcription of growth factors and their receptors / negative regulation of canonical NF-kappaB signal transduction / ESR-mediated signaling / negative regulation of miRNA transcription / TBP-class protein binding / nitric-oxide synthase regulator activity / nuclear estrogen receptor binding / transcription corepressor binding / transcription coregulator binding / stem cell differentiation / SUMOylation of intracellular receptors / cellular response to estradiol stimulus / euchromatin / beta-catenin binding / Nuclear Receptor transcription pathway / response to estrogen / transcription coactivator binding / male gonad development / nuclear receptor activity / positive regulation of fibroblast proliferation / Constitutive Signaling by Aberrant PI3K in Cancer / sequence-specific double-stranded DNA binding / positive regulation of nitric oxide biosynthetic process / Regulation of RUNX2 expression and activity / Ovarian tumor domain proteases / response to estradiol / PIP3 activates AKT signaling / positive regulation of cytosolic calcium ion concentration / ATPase binding / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / regulation of inflammatory response / DNA-binding transcription activator activity, RNA polymerase II-specific / fibroblast proliferation / transcription regulator complex / phospholipase C-activating G protein-coupled receptor signaling pathway / Estrogen-dependent gene expression / DNA-binding transcription factor activity, RNA polymerase II-specific / calmodulin binding / Extra-nuclear estrogen signaling / RNA polymerase II cis-regulatory region sequence-specific DNA binding / chromatin remodeling / DNA-binding transcription factor activity / negative regulation of gene expression / chromatin binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / protein kinase binding / chromatin / positive regulation of DNA-templated transcription / enzyme binding / negative regulation of transcription by RNA polymerase II / Golgi apparatus / signal transduction / positive regulation of transcription by RNA polymerase II / protein-containing complex / zinc ion binding / nucleoplasm / identical protein binding / nucleus / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Date: 2018 Dec Date: 2018 DecTitle: A Metastable Contact and Structural Disorder in the Estrogen Receptor Transactivation Domain Authors: Peng Y / Cao S / Kiselar J / Xiao X / Du Z / Hsieh A / Ko S / Chen Y / Agrawal P / Zheng W / Shi W / Jiang W / Yang L / Chance M / Surewicz W / Buck M |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDEE2 SASDEE2 |
|---|
-Related structure data
| Similar structure data |
|---|
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
| Model #2229 | 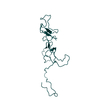 Type: atomic / Chi-square value: 0.136161 / P-value: 0.002500  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #2230 |  Type: atomic / Chi-square value: 0.341056 / P-value: 0.002500  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #2231 |  Type: atomic / Chi-square value: 0.509796 / P-value: 0.002500  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: The N-terminal domain of estrogen receptor alpha / Specimen concentration: 2.5 mg/ml |
|---|---|
| Buffer | Name: 20 mM sodium phosphate, 50 mM NaCl, 0.05 mM TCEP / pH: 7.4 |
| Entity #1227 | Name: ER-NTD / Type: protein Description: The N-terminal domain of estrogen receptor alpha Formula weight: 20.175 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: P03372 Sequence: SNAMTMTLHT KASGMALLHQ IQGNELEPLN RPQLKIPLER PLGEVYLDSS KPAVYNYPEG AAYEFNAAAA ANAQVYGQTG LPYGPGSEAA AFGSNGLGGF PPLNSVSPSP LMLLHPPPQL SPFLQPHGQQ VPYYLENEPS GYTVREAGPP AFYRPNSDNR RQGGRERLAS TNDKGSMAME SAKETRY |
-Experimental information
| Beam | Instrument name: National Synchrotron Light Source (NSLS-II) 16-ID (LiX) City: Brookhaven, NY / 国: USA  / Type of source: X-ray synchrotron / Wavelength: 0.09 Å / Dist. spec. to detc.: 3 mm / Type of source: X-ray synchrotron / Wavelength: 0.09 Å / Dist. spec. to detc.: 3 mm | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus3 S 1M / Pixsize x: 172 mm | |||||||||||||||||||||
| Scan |
| |||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller

















