+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | EspG3 chaperone from Mycobacterium smegmatis (Sel-Met labelled)
|
| Function / homology | EspG family / EspG family / cytoplasm / ESX-3 secretion-associated protein EspG3 Function and homology information Function and homology information |
| Biological species |  Mycobacterium smegmatis (bacteria) Mycobacterium smegmatis (bacteria) |
 Citation Citation |  Date: 2018 Jan 25 Date: 2018 Jan 25Title: Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems Authors: Tuukkanen A / Freire D / Chan S / Arbing M / Reed R / Evans T / Zenkeviciutė G / Kim J / Kahng S / Sawaya M / Wilmanns M / Eisenberg D / Parret A |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Models
- Sample
Sample
 Sample Sample | Name: EspG3 chaperone from Mycobacterium smegmatis (Sel-Met labelled) Specimen concentration: 3.79 mg/ml |
|---|---|
| Buffer | Name: 20 mM HEPES pH 7.5, 150 mM NaCl / pH: 7.5 |
| Entity #926 | Name: EspG3Msm / Type: protein / Description: EspG3 chaperone from Mycobacterium smegmatis / Formula weight: 31.8 / Num. of mol.: 1 / Source: Mycobacterium smegmatis / References: UniProt: A0QQ45 Sequence: GHMGPNAVEL TTDQAWCLAD VLGAGSYPWV LAITPPYSDH SQRSAFLAAQ SAELTRMGVV NSAGAVDPRV AQWITTVCRA TQWLDLRFVS GPGDLLRGMV ARRSEETVVA LRNAQLVTFT AMDIGHQHAL VPVLTAGLSG RKPARFDDFA LPAAAGARAD EQIRNGAPLA ...Sequence: GHMGPNAVEL TTDQAWCLAD VLGAGSYPWV LAITPPYSDH SQRSAFLAAQ SAELTRMGVV NSAGAVDPRV AQWITTVCRA TQWLDLRFVS GPGDLLRGMV ARRSEETVVA LRNAQLVTFT AMDIGHQHAL VPVLTAGLSG RKPARFDDFA LPAAAGARAD EQIRNGAPLA EVLEFLGVPP SARPLVESVF DGRRTYVEIV AGEHRDGHRV TTEVGVSIID TPHGRILVHP TKAFDGEWIS TFTPGSADAI AMAVERLTAS LPSGSWFPDQ PLTRDFDEDA ATHREPVLQR RTQKA |
-Experimental information
| Beam | Instrument name: PETRA III P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Type of source: X-ray synchrotron / Wavelength: 0.124 Å | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | |||||||||||||||||||||
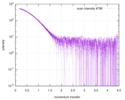
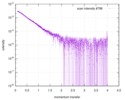
| Scan | Measurement date: Mar 17, 2014 / Exposure time: 0.05 sec. / Number of frames: 20 / Unit: 1/nm /
| |||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller



 SASDDU2
SASDDU2