+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | 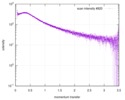 |
|---|---|
 試料 試料 | Carboyxamidomethylated ribonuclease A (unfolded RNAse) - with and without urea
|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報pancreatic ribonuclease / ribonuclease A activity / RNA nuclease activity / nucleic acid binding / defense response to Gram-positive bacterium / hydrolase activity / extracellular region 類似検索 - 分子機能 |
| 生物種 |  |
 引用 引用 |  ジャーナル: Biophys J / 年: 2018 ジャーナル: Biophys J / 年: 2018タイトル: Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions. 著者: Daniel Franke / Cy M Jeffries / Dmitri I Svergun /  要旨: Small-angle x-ray scattering (SAXS) of biological macromolecules in solutions is a widely employed method in structural biology. SAXS patterns include information about the overall shape and low- ...Small-angle x-ray scattering (SAXS) of biological macromolecules in solutions is a widely employed method in structural biology. SAXS patterns include information about the overall shape and low-resolution structure of dissolved particles. Here, we describe how to transform experimental SAXS patterns to feature vectors and how a simple k-nearest neighbor approach is able to retrieve information on overall particle shape and maximal diameter (D) as well as molecular mass directly from experimental scattering data. Based on this transformation, we develop a rapid multiclass shape-classification ranging from compact, extended, and flat categories to hollow and random-chain-like objects. This classification may be employed, e.g., as a decision block in automated data analysis pipelines. Further, we map protein structures from the Protein Data Bank into the classification space and, in a second step, use this mapping as a data source to obtain accurate estimates for the structural parameters (D, molecular mass) of the macromolecule under study based on the experimental scattering pattern alone, without inverse Fourier transform for D. All methods presented are implemented in a Fortran binary DATCLASS, part of the ATSAS data analysis suite, available on Linux, Mac, and Windows and free for academic use. |
 登録者 登録者 |
|
- 構造の表示
構造の表示
- ダウンロードとリンク
ダウンロードとリンク
-モデル
- 試料
試料
 試料 試料 | 名称: Carboyxamidomethylated ribonuclease A (unfolded RNAse) - with and without urea 試料濃度: 5.97 mg/ml |
|---|---|
| バッファ | 名称: 10 mM HCl / pH: 1 |
| 要素 #17 | 名称: RNase / タイプ: protein / 記述: Ribonuclease pancreatic / 分子量: 16.46 / 分子数: 1 / 由来: Bos taurus / 参照: UniProt: P61823 配列: MALKSLVLLS LLVLVLLLVR VQPSLGKETA AAKFERQHMD SSTSAASSSN YCNQMMKSRN LTKDRCKPVN TFVHESLADV QAVCSQKNVA CKNGQTNCYQ SYSTMSITDC RETGSSKYPN CAYKTTQANK HIIVACEGNP YVPVHFDASV |
-実験情報
| ビーム | 設備名称: PETRA III EMBL P12 / 地域: Hamburg / 国: Germany  / 線源: X-ray synchrotron / 波長: 0.124 Å / スペクトロメータ・検出器間距離: 3.1 mm / 線源: X-ray synchrotron / 波長: 0.124 Å / スペクトロメータ・検出器間距離: 3.1 mm | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 検出器 | 名称: Pilatus 2M | |||||||||||||||||||||||||||
| スキャン | 測定日: 2013年7月29日 / 保管温度: 10 °C / セル温度: 10 °C / 照射時間: 0.03 sec. / フレーム数: 17 / 単位: 1/nm /
| |||||||||||||||||||||||||||
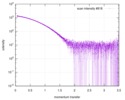
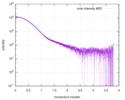
| 距離分布関数 P(R) |
| |||||||||||||||||||||||||||
| 結果 | コメント: Included in the additional files of the full-entry zip archive are SAXS data measured from folded RNAse (rnase.dat - also refer to SASBDB entry SASDDL3) as well as unfolded ...コメント: Included in the additional files of the full-entry zip archive are SAXS data measured from folded RNAse (rnase.dat - also refer to SASBDB entry SASDDL3) as well as unfolded carboyxamidomethylated ribonuclease A (rnaseU.dat) and unfolded RNAse in the presence of 1 M urea (rnaseUU1.dat) or 2 M urea (rnaseUU2.dat).
|
 ムービー
ムービー コントローラー
コントローラー



 SASDDM3
SASDDM3