[English] 日本語
 Yorodumi
Yorodumi- SASDDM3: Carboyxamidomethylated ribonuclease A (unfolded RNAse) - with and... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 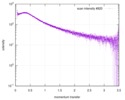 |
|---|---|
 Sample Sample | Carboyxamidomethylated ribonuclease A (unfolded RNAse) - with and without urea
|
| Function / homology |  Function and homology information Function and homology informationpancreatic ribonuclease / ribonuclease A activity / RNA nuclease activity / nucleic acid binding / defense response to Gram-positive bacterium / hydrolase activity / extracellular region Similarity search - Function |
| Biological species |  |
 Citation Citation |  Journal: Biophys J / Year: 2018 Journal: Biophys J / Year: 2018Title: Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions. Authors: Daniel Franke / Cy M Jeffries / Dmitri I Svergun /  Abstract: Small-angle x-ray scattering (SAXS) of biological macromolecules in solutions is a widely employed method in structural biology. SAXS patterns include information about the overall shape and low- ...Small-angle x-ray scattering (SAXS) of biological macromolecules in solutions is a widely employed method in structural biology. SAXS patterns include information about the overall shape and low-resolution structure of dissolved particles. Here, we describe how to transform experimental SAXS patterns to feature vectors and how a simple k-nearest neighbor approach is able to retrieve information on overall particle shape and maximal diameter (D) as well as molecular mass directly from experimental scattering data. Based on this transformation, we develop a rapid multiclass shape-classification ranging from compact, extended, and flat categories to hollow and random-chain-like objects. This classification may be employed, e.g., as a decision block in automated data analysis pipelines. Further, we map protein structures from the Protein Data Bank into the classification space and, in a second step, use this mapping as a data source to obtain accurate estimates for the structural parameters (D, molecular mass) of the macromolecule under study based on the experimental scattering pattern alone, without inverse Fourier transform for D. All methods presented are implemented in a Fortran binary DATCLASS, part of the ATSAS data analysis suite, available on Linux, Mac, and Windows and free for academic use. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDDM3 SASDDM3 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Carboyxamidomethylated ribonuclease A (unfolded RNAse) - with and without urea Specimen concentration: 5.97 mg/ml |
|---|---|
| Buffer | Name: 10 mM HCl / pH: 1 |
| Entity #17 | Name: RNase / Type: protein / Description: Ribonuclease pancreatic / Formula weight: 16.46 / Num. of mol.: 1 / Source: Bos taurus / References: UniProt: P61823 Sequence: MALKSLVLLS LLVLVLLLVR VQPSLGKETA AAKFERQHMD SSTSAASSSN YCNQMMKSRN LTKDRCKPVN TFVHESLADV QAVCSQKNVA CKNGQTNCYQ SYSTMSITDC RETGSSKYPN CAYKTTQANK HIIVACEGNP YVPVHFDASV |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | |||||||||||||||||||||||||||
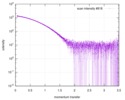
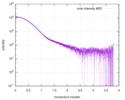
| Scan | Measurement date: Jul 29, 2013 / Storage temperature: 10 °C / Cell temperature: 10 °C / Exposure time: 0.03 sec. / Number of frames: 17 / Unit: 1/nm /
| |||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||
| Result | Comments: Included in the additional files of the full-entry zip archive are SAXS data measured from folded RNAse (rnase.dat - also refer to SASBDB entry SASDDL3) as well as unfolded ...Comments: Included in the additional files of the full-entry zip archive are SAXS data measured from folded RNAse (rnase.dat - also refer to SASBDB entry SASDDL3) as well as unfolded carboyxamidomethylated ribonuclease A (rnaseU.dat) and unfolded RNAse in the presence of 1 M urea (rnaseUU1.dat) or 2 M urea (rnaseUU2.dat).
|
 Movie
Movie Controller
Controller