+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDDB8 |
|---|---|
 Sample Sample | Periplasmic domain of inner membrane protein GspL, dimer
|
| Function / homology |  Function and homology information Function and homology informationGram-negative-bacterium-type cell wall / protein secretion by the type II secretion system / type II protein secretion system complex / plasma membrane Similarity search - Function |
| Biological species |  |
 Citation Citation |  Date: 2018 Dec Date: 2018 DecTitle: Structure and oligomerization of the periplasmic domain of GspL from the type II secretion system of Pseudomonas aeruginosa Authors: Fulara A / Vandenberghe I / Read R / Devreese B |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDDB8 SASDDB8 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
-Models
| Model #2072 |  Type: mix / Radius of dummy atoms: 1.90 A / Symmetry: none / Chi-square value: 5.788 / P-value: 0.000794  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #2073 |  Type: mix / Radius of dummy atoms: 1.90 A / Chi-square value: 5.788 / P-value: 0.000794  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #2074 |  Type: mix / Radius of dummy atoms: 1.90 A / Chi-square value: 5.788 / P-value: 0.000794  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #2075 |  Type: mix / Radius of dummy atoms: 1.90 A / Chi-square value: 5.788 / P-value: 0.000794  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #2076 |  Type: mix / Radius of dummy atoms: 1.90 A / Chi-square value: 5.788 / P-value: 0.000794  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #2077 |  Type: mix / Radius of dummy atoms: 1.90 A / Chi-square value: 5.788 / P-value: 0.000794  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Periplasmic domain of inner membrane protein GspL, dimer Specimen concentration: 2.00-13.70 |
|---|---|
| Buffer | Name: 50 mM TRIS, 100 mM NaCl / pH: 7.5 / Comment: pH=7.5 at 25°C |
| Entity #1111 | Name: GspLperi / Type: protein Description: Type II secretion system protein L, periplasmic domain Formula weight: 13.966 / Num. of mol.: 2 / Source: Pseudomonas aeruginosa / References: UniProt: P25060 Sequence: WSHPQFEKGG GDRYAAQSAE LYRQLFPEDR KLINLRAQFD QHLADSASSG GEGQLLGLLG QAATVIGGEP TVSVEQLDFS AARGDVALQV RAPGFDVLER LRSRLSESGL AVQLGSASRD GSTVSARLVI GG |
-Experimental information
| Beam | Instrument name: SOLEIL SWING  / City: Saint-Aubin / 国: France / City: Saint-Aubin / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.1033 Å / Type of source: X-ray synchrotron / Wavelength: 0.1033 Å | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: AVIEX PCCD170170 / Type: CCD | ||||||||||||||||||
| Scan | Measurement date: Apr 8, 2016 / Storage temperature: 4 °C / Cell temperature: 15 °C / Number of frames: 19 / Unit: 1/A /
| ||||||||||||||||||
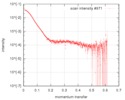
| Distance distribution function P(R) |
| ||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller