[English] 日本語
 Yorodumi
Yorodumi- SASDDB2: Phosphorylated Microtubule associated protein MAP2c (isoform 3); ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 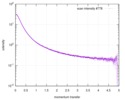 |
|---|---|
 Sample Sample | Phosphorylated Microtubule associated protein MAP2c (isoform 3); 6.8 mg/ml
|
| Function / homology | Isoform 3 of Microtubule-associated protein 2 Function and homology information Function and homology information |
| Biological species |  |
 Citation Citation |  Journal: J Biol Chem / Year: 2018 Journal: J Biol Chem / Year: 2018Title: Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics. Authors: Kateřina Melková / Vojtěch Zapletal / Séverine Jansen / Erik Nomilner / Milan Zachrdla / Jozef Hritz / Jiří Nováček / Markus Zweckstetter / Malene R Jensen / Martin Blackledge / Lukáš Žídek /    Abstract: Microtubule-associated protein 2c (MAP2c) is a 49-kDa intrinsically disordered protein regulating the dynamics of microtubules in developing neurons. MAP2c differs from its sequence homologue Tau in ...Microtubule-associated protein 2c (MAP2c) is a 49-kDa intrinsically disordered protein regulating the dynamics of microtubules in developing neurons. MAP2c differs from its sequence homologue Tau in the pattern and kinetics of phosphorylation by cAMP-dependent protein kinase (PKA). Moreover, the mechanisms through which MAP2c interacts with its binding partners and the conformational changes and dynamics associated with these interactions remain unclear. Here, we used NMR relaxation and paramagnetic relaxation enhancement techniques to determine the dynamics and long-range interactions within MAP2c. The relaxation rates revealed large differences in flexibility of individual regions of MAP2c, with the lowest flexibility observed in the known and proposed binding sites. Quantitative conformational analyses of chemical shifts, small-angle X-ray scattering (SAXS), and paramagnetic relaxation enhancement measurements disclosed that MAP2c regions interacting with important protein partners, including Fyn tyrosine kinase, plectin, and PKA, adopt specific conformations. High populations of polyproline II and α-helices were found in Fyn- and plectin-binding sites of MAP2c, respectively. The region binding the regulatory subunit of PKA consists of two helical motifs bridged by a more extended conformation. Of note, although MAP2c and Tau did not differ substantially in their conformations in regions of high sequence identity, we found that they differ significantly in long-range interactions, dynamics, and local conformation motifs in their N-terminal domains. These results highlight that the N-terminal regions of MAP2c provide important specificity to its regulatory roles and indicate a close relationship between MAP2c's biological functions and conformational behavior. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Models
- Sample
Sample
 Sample Sample | Name: Phosphorylated Microtubule associated protein MAP2c (isoform 3); 6.8 mg/ml Specimen concentration: 6.8 mg/ml |
|---|---|
| Buffer | Name: 50 mM MOPS, 150 mM NaCl, 0.03% NaN3 / pH: 6.9 / Comment: aka: MOPS map2c buffer |
| Entity #917 | Name: MAP2c / Type: protein / Description: Microtubule-associated protein 2, isoform 3 / Formula weight: 49.299 / Num. of mol.: 1 / Source: Rattus norvegicus / References: UniProt: P15146-3 Sequence: MADERKDEGK APHWTSASLT EAAAHPHSPE MKDQGGSGEG LSRSANGFPY REEEEGAFGE HGSQGTYSDT KENGINGELT SADRETAEEV SARIVQVVTA EAVAVLKGEQ EKEAQHKDQP AALPLAAEET VNLPPSPPPS PASEQTAALE EATSGESAQA PSAFKQAKDK ...Sequence: MADERKDEGK APHWTSASLT EAAAHPHSPE MKDQGGSGEG LSRSANGFPY REEEEGAFGE HGSQGTYSDT KENGINGELT SADRETAEEV SARIVQVVTA EAVAVLKGEQ EKEAQHKDQP AALPLAAEET VNLPPSPPPS PASEQTAALE EATSGESAQA PSAFKQAKDK VTDGITKSPE KRSSLPRPSS ILPPRRGVSG DREENSFSLN SSISSARRTT RSEPIRRAGK SGTSTPTTPG STAITPGTPP SYSSRTPGTP GTPSYPRTPG TPKSGILVPS EKKVAIIRTP PKSPATPKQL RLINQPLPDL KNVKSKIGST DNIKYQPKGG QVQIVTKKID LSHVTSKCGS LKNIRHRPGG GRVKIESVKL DFKEKAQAKV GSLDNAHHVP GGGNVKIDSQ KLNFREHAKA RVDHGAEIIT QSPSRSSVAS PRRLSNVSSS GSINLLESPQ LATLAEDVTA ALAKQGL |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.099 Å / Type of source: X-ray synchrotron / Wavelength: 0.099 Å | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | |||||||||||||||
| Scan | Measurement date: Apr 21, 2017 / Storage temperature: 20 °C / Cell temperature: 20 °C / Exposure time: 1 sec. / Number of frames: 10 / Unit: 1/nm /
| |||||||||||||||
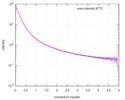
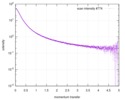
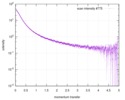
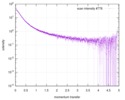
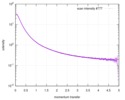
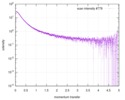
| Result | Experimental MW: 66 kDa / Type of curve: single_conc
|
 Movie
Movie Controller
Controller


 SASDDB2
SASDDB2