+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | Rab family protein CtRoco L487A GDP
|
| Function / homology |  Function and homology information Function and homology informationnon-specific serine/threonine protein kinase / protein serine/threonine kinase activity / GTP binding / ATP binding / identical protein binding Similarity search - Function |
| Biological species |  Chlorobium tepidum (strain ATCC 49652 / DSM 12025 / NBRC 103806 / TLS) (bacteria) Chlorobium tepidum (strain ATCC 49652 / DSM 12025 / NBRC 103806 / TLS) (bacteria) |
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover. Authors: Egon Deyaert / Lina Wauters / Giambattista Guaitoli / Albert Konijnenberg / Margaux Leemans / Susanne Terheyden / Arsen Petrovic / Rodrigo Gallardo / Laura M Nederveen-Schippers / Panagiotis ...Authors: Egon Deyaert / Lina Wauters / Giambattista Guaitoli / Albert Konijnenberg / Margaux Leemans / Susanne Terheyden / Arsen Petrovic / Rodrigo Gallardo / Laura M Nederveen-Schippers / Panagiotis S Athanasopoulos / Henderikus Pots / Peter J M Van Haastert / Frank Sobott / Christian Johannes Gloeckner / Rouslan Efremov / Arjan Kortholt / Wim Versées /     Abstract: Mutations in LRRK2 are a common cause of genetic Parkinson's disease (PD). LRRK2 is a multi-domain Roco protein, harbouring kinase and GTPase activity. In analogy with a bacterial homologue, LRRK2 ...Mutations in LRRK2 are a common cause of genetic Parkinson's disease (PD). LRRK2 is a multi-domain Roco protein, harbouring kinase and GTPase activity. In analogy with a bacterial homologue, LRRK2 was proposed to act as a GTPase activated by dimerization (GAD), while recent reports suggest LRRK2 to exist under a monomeric and dimeric form in vivo. It is however unknown how LRRK2 oligomerization is regulated. Here, we show that oligomerization of a homologous bacterial Roco protein depends on the nucleotide load. The protein is mainly dimeric in the nucleotide-free and GDP-bound states, while it forms monomers upon GTP binding, leading to a monomer-dimer cycle during GTP hydrolysis. An analogue of a PD-associated mutation stabilizes the dimer and decreases the GTPase activity. This work thus provides insights into the conformational cycle of Roco proteins and suggests a link between oligomerization and disease-associated mutations in LRRK2. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDCE2 SASDCE2 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Rab family protein CtRoco L487A GDP / Specimen concentration: 1.00-2.00 |
|---|---|
| Buffer | Name: 20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT 200µM GDP pH: 7.5 |
| Entity #601 | Name: CtRoco / Type: protein / Description: Rab family protein / Formula weight: 127.147 / Num. of mol.: 2 Source: Chlorobium tepidum (strain ATCC 49652 / DSM 12025 / NBRC 103806 / TLS) References: UniProt: Q8KC98 Sequence: MSYYHHHHHH DYDIPTTENL YFQGAMGSMS DLDVIRQIEQ ELGMQLEPVD KLKWYSKGYK LDKDQRVTAI GLYDCGSDTL DRIIQPLESL KSLSELSLSS NQITDISPLA SLNSLSMLWL DRNQITDIAP LASLNSLSML WLFGNKISDI APLESLKSLT ELQLSSNQIT ...Sequence: MSYYHHHHHH DYDIPTTENL YFQGAMGSMS DLDVIRQIEQ ELGMQLEPVD KLKWYSKGYK LDKDQRVTAI GLYDCGSDTL DRIIQPLESL KSLSELSLSS NQITDISPLA SLNSLSMLWL DRNQITDIAP LASLNSLSML WLFGNKISDI APLESLKSLT ELQLSSNQIT DIAPLASLKS LTELSLSGNN ISDIAPLESL KSLTELSLSS NQITDIAPLA SLKSLTELSL SSNQISDIAP LESLKSLTEL QLSRNQISDI APLESLKSLT ELQLSSNQIT DIAPLASLKS LTELQLSRNQ ISDIAPLESL NSLSKLWLNG NQITDIAPLA SLNSLTELEL SSNQITDIAP LASLKSLSTL WLSSNQISDI APLASLESLS ELSLSSNQIS DISPLASLNS LTGFDVRRNP IKRLPETITG FDMEILWNDF SSSGFITFFD NPLESPPPEI VKQGKEAVRQ YFQSIEEARS KGEALVHLQE IKVHLIGDGM AGKTSLLKQL IGETFDPKES QTHGLNVVTK QAPNIKGLEN DDELKECLFH FWDFGGQEIM HASHQFFMTR SSVYMLLLDS RTDSNKHYWL RHIEKYGGKS PVIVVMNKID ENPSYNIEQK KINERFPAIE NRFHRISCKN GDGVESIAKS LKSAVLHPDS IYGTPLAPSW IKVKEKLVEA TTAQRYLNRT EVEKICNDSG ITDPGERKTL LGYLNNLGIV LYFEALDLSE IYVLDPHWVT IGVYRIINSS KTKNGHLNTS ALGYILNEEQ IRCDEYDPAK NNKFTYTLLE QRYLLDIMKQ FELCYDEGKG LFIIPSNLPT QIDNEPEITE GEPLRFIMKY DYLPSTIIPR LMIAMQHQIL DRMQWRYGMV LKSQDHEGAL AKVVAETKDS TITIAIQGEP RCKREYLSII WYEIKKINAN FTNLDVKEFI PLPGHPDELV EYKELLGLEK MGRDEYVSGK LEKVFSVSKM LDSVISKEER NKERLMGDIN IKLENIGNPT IPIHQQVEVN VSQETVQHVE NLQGFFENLK ADILREAELE IDDPKERKRL ANELELAENA ITKMDAAVKS GKNKLKPDVK DRLGEFIDNL ANENSRLRKG IALVMNGAEK VQKLARYYNN VAPFFDLPSV PPVLLGKEKT |
-Experimental information
| Beam | Instrument name: SOLEIL SWING  / City: Saint-Aubin / 国: France / City: Saint-Aubin / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.103 Å / Type of source: X-ray synchrotron / Wavelength: 0.103 Å | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: AVIEX / Type: CCD | |||||||||||||||||||||||||||||||||
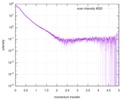
| Scan |
| |||||||||||||||||||||||||||||||||
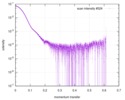
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
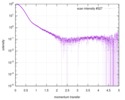
| Result |
|
 Movie
Movie Controller
Controller