+検索条件
-Structure paper
| タイトル | A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover. |
|---|---|
| ジャーナル・号・ページ | Nat Commun, Vol. 8, Issue 1, Page 1008, Year 2017 |
| 掲載日 | 2017年10月18日 |
 著者 著者 | Egon Deyaert / Lina Wauters / Giambattista Guaitoli / Albert Konijnenberg / Margaux Leemans / Susanne Terheyden / Arsen Petrovic / Rodrigo Gallardo / Laura M Nederveen-Schippers / Panagiotis S Athanasopoulos / Henderikus Pots / Peter J M Van Haastert / Frank Sobott / Christian Johannes Gloeckner / Rouslan Efremov / Arjan Kortholt / Wim Versées /     |
| PubMed 要旨 | Mutations in LRRK2 are a common cause of genetic Parkinson's disease (PD). LRRK2 is a multi-domain Roco protein, harbouring kinase and GTPase activity. In analogy with a bacterial homologue, LRRK2 ...Mutations in LRRK2 are a common cause of genetic Parkinson's disease (PD). LRRK2 is a multi-domain Roco protein, harbouring kinase and GTPase activity. In analogy with a bacterial homologue, LRRK2 was proposed to act as a GTPase activated by dimerization (GAD), while recent reports suggest LRRK2 to exist under a monomeric and dimeric form in vivo. It is however unknown how LRRK2 oligomerization is regulated. Here, we show that oligomerization of a homologous bacterial Roco protein depends on the nucleotide load. The protein is mainly dimeric in the nucleotide-free and GDP-bound states, while it forms monomers upon GTP binding, leading to a monomer-dimer cycle during GTP hydrolysis. An analogue of a PD-associated mutation stabilizes the dimer and decreases the GTPase activity. This work thus provides insights into the conformational cycle of Roco proteins and suggests a link between oligomerization and disease-associated mutations in LRRK2. |
 リンク リンク |  Nat Commun / Nat Commun /  PubMed:29044096 / PubMed:29044096 /  PubMed Central PubMed Central |
| 手法 | SAS (X-ray synchrotron) |
| 構造データ |  SASDC82: 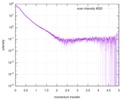 SASDC92: Rab family protein CtRoco GppNHp (Rab family protein, CtRoco)  SASDCA2: Rab family protein CtRoco GDP (Rab family protein, CtRoco)  SASDCB2:  SASDCC2: Rab family protein CtRoc-COR GDP (Rab family protein) 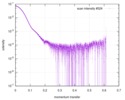 SASDCD2: Rab family protein CtRoc-COR GppNHp (Rab family protein)  SASDCE2: Rab family protein CtRoco L487A GDP (Rab family protein, CtRoco)  SASDCF2: Rab family protein CtRoco L487A GppNHp (Rab family protein, CtRoco) 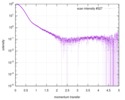 SASDCG2: Rab family protein CtRoco L487A (Rab family protein, CtRoco) |
| 由来 |
|
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア 万見文献について
万見文献について



 Chlorobium tepidum (strain atcc 49652 / dsm 12025 / nbrc 103806 / tls) (バクテリア)
Chlorobium tepidum (strain atcc 49652 / dsm 12025 / nbrc 103806 / tls) (バクテリア)