[English] 日本語
 Yorodumi
Yorodumi- SASDBS5: Cyclohexanone monooxygenase, N497A-K501A (Cyclohexanone monooxyge... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | Cyclohexanone monooxygenase, N497A-K501A
|
| Function / homology | : / Flavin monooxygenase-like / Flavin-binding monooxygenase-like / N,N-dimethylaniline monooxygenase activity / FAD/NAD(P)-binding domain superfamily / NADP binding / flavin adenine dinucleotide binding / Cyclohexanone monooxygenase Function and homology information Function and homology information |
| Biological species |  Rhodococcus sp. HI-31 (bacteria) Rhodococcus sp. HI-31 (bacteria) |
 Citation Citation |  Journal: Biochim Biophys Acta / Year: 2016 Journal: Biochim Biophys Acta / Year: 2016Title: The role of conformational flexibility in Baeyer-Villiger monooxygenase catalysis and structure. Authors: Brahm J Yachnin / Peter C K Lau / Albert M Berghuis /  Abstract: BACKGROUND: The Baeyer-Villiger monooxygenases (BMVOs) are a group of microbial enzymes that have garnered interest as industrial biocatalysts. While great strides have been made in recent years to ...BACKGROUND: The Baeyer-Villiger monooxygenases (BMVOs) are a group of microbial enzymes that have garnered interest as industrial biocatalysts. While great strides have been made in recent years to understand the mechanism of these enzymes from a structural perspective, our understanding remains incomplete. In particular, the role of a twenty residue loop (residues 487-504), which we refer to as the "Control Loop," that is observed in either an ordered or disordered state in various crystal structures remains unclear. METHODS: Using SAXS, we have made the first observations of the Loop in solution with two BVMOs, cyclohexanone monooxygenase (CHMO) and cyclopentadecanone monooxygenase. We also made a series of ...METHODS: Using SAXS, we have made the first observations of the Loop in solution with two BVMOs, cyclohexanone monooxygenase (CHMO) and cyclopentadecanone monooxygenase. We also made a series of mutants of CHMO and analyzed them using SAXS, ITC, and an uncoupling assay. RESULTS: These experiments show that Control Loop ordering results in an overall more compact enzyme without altering global protein foldedness. We have also demonstrated that the Loop plays a ...RESULTS: These experiments show that Control Loop ordering results in an overall more compact enzyme without altering global protein foldedness. We have also demonstrated that the Loop plays a critical and complex role on enzyme structure and catalysis. The Control Loop appears to have a direct impact on the organization of the overall structure of the protein, as well as in influencing the active site environment. CONCLUSIONS: The data imply that the Loop can be divided into two regions, referred to as "sub-loops," that coordinate overall domain movements to changes in the active site. GENERAL SIGNIFICANCE: A better understanding of the mechanistic role of the Control Loop may ultimately be helpful in designing mutants with altered specificity and improved catalytic efficiency. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Models
- Sample
Sample
 Sample Sample | Name: Cyclohexanone monooxygenase, N497A-K501A / Specimen concentration: 2.00-8.00 |
|---|---|
| Buffer | Name: 50 mM Tris 5 mM NADP+ / pH: 8 |
| Entity #365 | Name: CHMO-K501A / Type: protein / Description: Cyclohexanone monooxygenase / Formula weight: 60.689 / Num. of mol.: 1 / Source: Rhodococcus sp. HI-31 / References: UniProt: C0STX7 Sequence: GSLEASMHMT AQTTHTVDAV VIGAGFGGIY AVHKLHHELG LTTVGFDKAD GPGGTWYWNR YPGALSDTES HLYRFSFDRD LLQESTWKTT YITQPEILEY LEDVVDRFDL RRHFKFGTEV TSALYLDDEN LWEVTTDHGE VYRAKYVVNA VGLLSAINFP NLPGLDTFEG ...Sequence: GSLEASMHMT AQTTHTVDAV VIGAGFGGIY AVHKLHHELG LTTVGFDKAD GPGGTWYWNR YPGALSDTES HLYRFSFDRD LLQESTWKTT YITQPEILEY LEDVVDRFDL RRHFKFGTEV TSALYLDDEN LWEVTTDHGE VYRAKYVVNA VGLLSAINFP NLPGLDTFEG ETIHTAAWPE GKSLAGRRVG VIGTGSTGQQ VITSLAPEVE HLTVFVRTPQ YSVPVGNRPV NPEQIAEIKA DYDRIWERAK NSAVAFGFEE STLPAMSVSE EERNRIFQEA WDHGGGFRFM FGTFGDIATD EAANEAAASF IRAKVAEIIE DPETARKLMP KGLFAKRPLC DSGYYEVYNR PNVEAVAIKE NPIREVTAKG VVTEDGVLHE LDVLVFATGF DAVDGNYRRI EIRGRDGLHI NDHWDGQPTS YLGVSTANFP NWFMVLGPNG PFTNLPPSIE TQVEWISDTI GYAERNGVRA IEPTPEAEAE WTETCTEIAN ATLFTKGDSW IFGANIPGAK PSVLFYLGGL RNYRAVMAEV AADGYRGFEV KSAEMVTV |
-Experimental information
| Beam | Instrument name: Advanced Light Source (ALS) 12.3.1 (SIBYLS) City: Berkeley, CA / 国: USA  / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 1.5 mm / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 1.5 mm | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: MAR 165 CCD | |||||||||||||||||||||
| Scan |
| |||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||
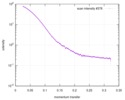
| Result |
|
 Movie
Movie Controller
Controller


 SASDBS5
SASDBS5