+ Open data
Open data
- Basic information
Basic information
| Entry | 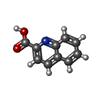 Database: PDB chemical components / ID: QNC Database: PDB chemical components / ID: QNC |
|---|---|
| Name | Name: |
-Chemical information
| Composition |  | ||||
|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAIN / Three letter code: QNC / Ideal coordinates details: Corina / Model coordinates PDB-ID: 1JLD | ||||
| History |
| ||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Brenda Brenda   / /  ChEBI / ChEBI /  ChemicalBook / ChemicalBook /  CompTox / CompTox /  DrugBank / DrugBank /  HMDB / HMDB /  KEGG_Ligand / KEGG_Ligand /  Metabolights / Metabolights /  Nikkaji / Nikkaji /  PubChem / PubChem /  PubChem_TPharma / PubChem_TPharma /  SureChEMBL / SureChEMBL /  ZINC / ZINC /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 12.01 | | CACTVS 3.370 | OpenEye OEToolkits 1.7.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.370 | | OpenEye OEToolkits 1.7.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 12.01 | | OpenEye OEToolkits 1.7.0 | |
|---|
-PDB entries
Showing all 5 items

PDB-2fgu: 
X-ray crystal structure of HIV-1 Protease T80S variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity.
PDB-2fgv: 
X-ray crystal structure of HIV-1 Protease T80N variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity.

PDB-4jku: 
Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinaldic acid, NYSGRC Target 14306

PDB-7x6j: 
SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3af

PDB-9dl3: 
Structure of proline utilization A complexed with quinoline-2-carboxylic acid
 Movie
Movie Controller
Controller




