[English] 日本語
 Yorodumi
Yorodumi- PDB-8pm2: Structure of the murine trace amine-associated receptor TAAR7f bo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8pm2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | MEMBRANE PROTEIN / trace-amine associated receptor / TAAR / mTAAR7f / GPCR / receptor / G protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationtrace-amine receptor activity / sensory perception of chemical stimulus / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels ...trace-amine receptor activity / sensory perception of chemical stimulus / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / retina development in camera-type eye / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Gusach, A. / Lee, Y. / Edwards, P.C. / Huang, F. / Weyand, S.N. / Tate, C.G. | |||||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f. Authors: Anastasiia Gusach / Yang Lee / Armin Nikpour Khoshgrudi / Elizaveta Mukhaleva / Ning Ma / Eline J Koers / Qingchao Chen / Patricia C Edwards / Fanglu Huang / Jonathan Kim / Filippo Mancia / ...Authors: Anastasiia Gusach / Yang Lee / Armin Nikpour Khoshgrudi / Elizaveta Mukhaleva / Ning Ma / Eline J Koers / Qingchao Chen / Patricia C Edwards / Fanglu Huang / Jonathan Kim / Filippo Mancia / Dmitry B Verprintsev / Nagarajan Vaidehi / Simone N Weyand / Christopher G Tate /   Abstract: There are two main families of G protein-coupled receptors that detect odours in humans, the odorant receptors (ORs) and the trace amine-associated receptors (TAARs). Their amino acid sequences are ...There are two main families of G protein-coupled receptors that detect odours in humans, the odorant receptors (ORs) and the trace amine-associated receptors (TAARs). Their amino acid sequences are distinct, with the TAARs being most similar to the aminergic receptors such as those activated by adrenaline, serotonin and histamine. To elucidate the structural determinants of ligand recognition by TAARs, we have determined the cryo-EM structure of a murine receptor, mTAAR7f, coupled to the heterotrimeric G protein G and bound to the odorant N,N-dimethylcyclohexylamine (DMCH) to an overall resolution of 2.9 Å. DMCH is bound in a hydrophobic orthosteric binding site primarily through van der Waals interactions and a strong charge-charge interaction between the tertiary amine of the ligand and an aspartic acid residue. This site is distinct and non-overlapping with the binding site for the odorant propionate in the odorant receptor OR51E2. The structure, in combination with mutagenesis data and molecular dynamics simulations suggests that the activation of the receptor follows a similar pathway to that of the β-adrenoceptors, with the significant difference that DMCH interacts directly with one of the main activation microswitch residues. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8pm2.cif.gz 8pm2.cif.gz | 215 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8pm2.ent.gz pdb8pm2.ent.gz | 160.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8pm2.json.gz 8pm2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  8pm2_validation.pdf.gz 8pm2_validation.pdf.gz | 1.3 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  8pm2_full_validation.pdf.gz 8pm2_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  8pm2_validation.xml.gz 8pm2_validation.xml.gz | 47.8 KB | Display | |
| Data in CIF |  8pm2_validation.cif.gz 8pm2_validation.cif.gz | 70.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pm/8pm2 https://data.pdbj.org/pub/pdb/validation_reports/pm/8pm2 ftp://data.pdbj.org/pub/pdb/validation_reports/pm/8pm2 ftp://data.pdbj.org/pub/pdb/validation_reports/pm/8pm2 | HTTPS FTP |
-Related structure data
| Related structure data |  17756MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules ABG
| #1: Protein | Mass: 28964.734 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNAS, GNAS1, GSP / Plasmid: pET15b / Production host: Homo sapiens (human) / Gene: GNAS, GNAS1, GSP / Plasmid: pET15b / Production host:  |
|---|---|
| #2: Protein | Mass: 37342.785 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNB1 / Production host: Homo sapiens (human) / Gene: GNB1 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P62873 Trichoplusia ni (cabbage looper) / References: UniProt: P62873 |
| #3: Protein | Mass: 7845.078 Da / Num. of mol.: 1 / Mutation: C68S Source method: isolated from a genetically manipulated source Details: There is an engineered mutation C68S introduced / Source: (gene. exp.)  Homo sapiens (human) / Gene: GNG2 / Production host: Homo sapiens (human) / Gene: GNG2 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P59768 Trichoplusia ni (cabbage looper) / References: UniProt: P59768 |
-Antibody / Protein , 2 types, 2 molecules NR
| #4: Antibody | Mass: 16926.076 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #5: Protein | Mass: 41012.539 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: mTAAR7f sequence contains cleaved protease sites: TEV (S residue on the N terminus) and HRV-3C (C terminus) Source: (gene. exp.)   Trichoplusia ni (cabbage looper) / References: UniProt: Q5QD08 Trichoplusia ni (cabbage looper) / References: UniProt: Q5QD08 |
-Non-polymers , 2 types, 2 molecules 
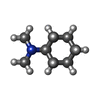

| #6: Chemical | ChemComp-Y01 / |
|---|---|
| #7: Chemical | ChemComp-8IA / ~{ |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit ...Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit alpha isoform short + Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 + Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 + Nanobody 35 Type: COMPLEX / Entity ID: #1-#5 / Source: RECOMBINANT | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||||||||||||
| Source (recombinant) | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) | ||||||||||||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||||||||||||||
| Specimen | Conc.: 0.8 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||||||||||||||||||
| Specimen support | Details: Forward Power of 38 W, Reflected Power of 2W; Fischione Grid material: GOLD / Grid mesh size: 400 divisions/in. / Grid type: UltrAuFoil R1.2/1.3 | ||||||||||||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 96000 X / Calibrated magnification: 96000 X / Nominal defocus max: 2400 nm / Nominal defocus min: 800 nm / Calibrated defocus min: 2400 nm / Calibrated defocus max: 2400 nm / Cs: 2.7 mm / C2 aperture diameter: 100 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 7.84 sec. / Electron dose: 55 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 11157 |
| Image scans | Width: 4096 / Height: 4096 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: Done with cryoSPARC patch motion correction / Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 9199357 / Details: Particles were heavily over-picked | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.92 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 172639 / Algorithm: FOURIER SPACE / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: OTHER / Space: REAL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Details: Initial models of heterotrimeric mini-Gs and Nb35 were sourced from PDB 7T9I. A de novo model of TAAR7f was generated from the focused map and protein sequence using ModelAngelo Source name: Other / Type: other | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 2.92→2.92 Å / Cor.coef. Fo:Fc: 0.836 / SU B: 13.865 / SU ML: 0.249 / ESU R: 0.346 Stereochemistry target values: MAXIMUM LIKELIHOOD WITH PHASES Details: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: PARAMETERS FOR MASK CACLULATION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 104.792 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Total: 7624 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj























