+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8gus | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HU-CB2-G protein complex | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | STRUCTURAL PROTEIN / GPCR / G protein / cryo-EM / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationcannabinoid receptor activity / negative regulation of mast cell activation / negative regulation of synaptic transmission, GABAergic / negative regulation of action potential / Class A/1 (Rhodopsin-like receptors) / regulation of metabolic process / leukocyte chemotaxis / extrinsic component of cytoplasmic side of plasma membrane / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / T cell migration ...cannabinoid receptor activity / negative regulation of mast cell activation / negative regulation of synaptic transmission, GABAergic / negative regulation of action potential / Class A/1 (Rhodopsin-like receptors) / regulation of metabolic process / leukocyte chemotaxis / extrinsic component of cytoplasmic side of plasma membrane / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / T cell migration / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / regulation of mitotic spindle organization / cellular response to forskolin / response to amphetamine / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / response to peptide hormone / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / GDP binding / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / retina development in camera-type eye / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / cell cortex / midbody / G alpha (i) signalling events / fibroblast proliferation / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / G alpha (s) signalling events / G alpha (q) signalling events / perikaryon / Ras protein signal transduction / response to lipopolysaccharide / cell population proliferation / Extra-nuclear estrogen signaling / inflammatory response / immune response / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / centrosome / dendrite / synapse / protein-containing complex binding / nucleolus / GTP binding / magnesium ion binding / signal transduction / endoplasmic reticulum / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.97 Å | |||||||||
 Authors Authors | Wu, L.J. / Hua, T. / Liu, Z.J. / Li, X.T. / Chang, H. | |||||||||
| Funding support |  China, 2items China, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of selective cannabinoid CB receptor activation. Authors: Xiaoting Li / Hao Chang / Jara Bouma / Laura V de Paus / Partha Mukhopadhyay / Janos Paloczi / Mohammed Mustafa / Cas van der Horst / Sanjay Sunil Kumar / Lijie Wu / Yanan Yu / Richard J B H ...Authors: Xiaoting Li / Hao Chang / Jara Bouma / Laura V de Paus / Partha Mukhopadhyay / Janos Paloczi / Mohammed Mustafa / Cas van der Horst / Sanjay Sunil Kumar / Lijie Wu / Yanan Yu / Richard J B H N van den Berg / Antonius P A Janssen / Aron Lichtman / Zhi-Jie Liu / Pal Pacher / Mario van der Stelt / Laura H Heitman / Tian Hua /    Abstract: Cannabinoid CB receptor (CBR) agonists are investigated as therapeutic agents in the clinic. However, their molecular mode-of-action is not fully understood. Here, we report the discovery of LEI-102, ...Cannabinoid CB receptor (CBR) agonists are investigated as therapeutic agents in the clinic. However, their molecular mode-of-action is not fully understood. Here, we report the discovery of LEI-102, a CBR agonist, used in conjunction with three other CBR ligands (APD371, HU308, and CP55,940) to investigate the selective CBR activation by binding kinetics, site-directed mutagenesis, and cryo-EM studies. We identify key residues for CBR activation. Highly lipophilic HU308 and the endocannabinoids, but not the more polar LEI-102, APD371, and CP55,940, reach the binding pocket through a membrane channel in TM1-TM7. Favorable physico-chemical properties of LEI-102 enable oral efficacy in a chemotherapy-induced nephropathy model. This study delineates the molecular mechanism of CBR activation by selective agonists and highlights the role of lipophilicity in CBR engagement. This may have implications for GPCR drug design and sheds light on their activation by endogenous ligands. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8gus.cif.gz 8gus.cif.gz | 235.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8gus.ent.gz pdb8gus.ent.gz | 182.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8gus.json.gz 8gus.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  8gus_validation.pdf.gz 8gus_validation.pdf.gz | 1.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  8gus_full_validation.pdf.gz 8gus_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  8gus_validation.xml.gz 8gus_validation.xml.gz | 42.4 KB | Display | |
| Data in CIF |  8gus_validation.cif.gz 8gus_validation.cif.gz | 62.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gu/8gus https://data.pdbj.org/pub/pdb/validation_reports/gu/8gus ftp://data.pdbj.org/pub/pdb/validation_reports/gu/8gus ftp://data.pdbj.org/pub/pdb/validation_reports/gu/8gus | HTTPS FTP |
-Related structure data
| Related structure data |  34278MC  8guqC  8gurC  8gutC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules ABC
| #1: Protein | Mass: 40415.031 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNAI1 / Production host: Homo sapiens (human) / Gene: GNAI1 / Production host:  |
|---|---|
| #2: Protein | Mass: 37416.930 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNB1 / Production host: Homo sapiens (human) / Gene: GNB1 / Production host:  |
| #3: Protein | Mass: 7861.143 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNG2 / Production host: Homo sapiens (human) / Gene: GNG2 / Production host:  |
-Protein / Antibody / Non-polymers , 3 types, 3 molecules RS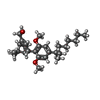

| #4: Protein | Mass: 39722.715 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CNR2, CB2A, CB2B / Production host: Homo sapiens (human) / Gene: CNR2, CB2A, CB2B / Production host:  |
|---|---|
| #5: Antibody | Mass: 27784.896 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  |
| #6: Chemical | ChemComp-KO3 / [( |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Ternary complex of Cannabinoid receptor 2 with Guanine nucleotide-binding protein and Single-chain variable fragment Type: COMPLEX / Entity ID: #1-#5 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 140 kDa/nm / Experimental value: YES |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: GOLD / Grid mesh size: 400 divisions/in. / Grid type: Quantifoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 105000 X / Nominal defocus max: 2000 nm / Nominal defocus min: 1000 nm / Cs: 2.7 mm / C2 aperture diameter: 50 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Temperature (max): 70 K / Temperature (min): 70 K |
| Image recording | Electron dose: 1.5 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
| EM imaging optics | Energyfilter name: GIF Quantum LS / Energyfilter slit width: 20 eV |
- Processing
Processing
| EM software |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | |||||||||
| 3D reconstruction | Resolution: 2.97 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 355832 / Algorithm: FOURIER SPACE / Symmetry type: POINT | |||||||||
| Atomic model building | B value: 68.91 / Protocol: RIGID BODY FIT / Space: REAL | |||||||||
| Atomic model building | PDB-ID: 6KPF Accession code: 6KPF / Source name: PDB / Type: experimental model |
 Movie
Movie Controller
Controller






 PDBj
PDBj































