[English] 日本語
 Yorodumi
Yorodumi- PDB-8g3h: Structure of cobalamin-dependent methionine synthase (MetH) in a ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8g3h | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
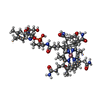
| Title | Structure of cobalamin-dependent methionine synthase (MetH) in a resting state | |||||||||
 Components Components | Methionine synthase | |||||||||
 Keywords Keywords | TRANSFERASE / Methyltransferase / Amino-acid biosynthesis / Methionine biosynthesis | |||||||||
| Function / homology |  Function and homology information Function and homology informationmethionine synthase / methionine synthase activity / homocysteine metabolic process / cobalamin binding / tetrahydrofolate metabolic process / methylation / zinc ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |   Thermus filiformis (bacteria) Thermus filiformis (bacteria) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Watkins, M.B. / Ando, N. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Conformational switching and flexibility in cobalamin-dependent methionine synthase studied by small-angle X-ray scattering and cryoelectron microscopy. Authors: Maxwell B Watkins / Haoyue Wang / Audrey Burnim / Nozomi Ando /  Abstract: Cobalamin-dependent methionine synthase (MetH) catalyzes the synthesis of methionine from homocysteine and 5-methyltetrahydrofolate (CH-Hfolate) using the unique chemistry of its cofactor. In doing ...Cobalamin-dependent methionine synthase (MetH) catalyzes the synthesis of methionine from homocysteine and 5-methyltetrahydrofolate (CH-Hfolate) using the unique chemistry of its cofactor. In doing so, MetH links the cycling of -adenosylmethionine with the folate cycle in one-carbon metabolism. Extensive biochemical and structural studies on MetH have shown that this flexible, multidomain enzyme adopts two major conformations to prevent a futile cycle of methionine production and consumption. However, as MetH is highly dynamic as well as both a photosensitive and oxygen-sensitive metalloenzyme, it poses special challenges for structural studies, and existing structures have necessarily come from a "divide and conquer" approach. In this study, we investigate MetH and a thermophilic homolog from using small-angle X-ray scattering (SAXS), single-particle cryoelectron microscopy (cryo-EM), and extensive analysis of the AlphaFold2 database to present a structural description of the full-length MetH in its entirety. Using SAXS, we describe a common resting-state conformation shared by both active and inactive oxidation states of MetH and the roles of CH-Hfolate and flavodoxin in initiating turnover and reactivation. By combining SAXS with a 3.6-Å cryo-EM structure of the MetH, we show that the resting-state conformation consists of a stable arrangement of the catalytic domains that is linked to a highly mobile reactivation domain. Finally, by combining AlphaFold2-guided sequence analysis and our experimental findings, we propose a general model for functional switching in MetH. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8g3h.cif.gz 8g3h.cif.gz | 165.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8g3h.ent.gz pdb8g3h.ent.gz | 123.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8g3h.json.gz 8g3h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g3/8g3h https://data.pdbj.org/pub/pdb/validation_reports/g3/8g3h ftp://data.pdbj.org/pub/pdb/validation_reports/g3/8g3h ftp://data.pdbj.org/pub/pdb/validation_reports/g3/8g3h | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  29699MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 131312.266 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Thermus filiformis (bacteria) / Gene: THFILI_06775 / Plasmid: pET-28c+ / Production host: Thermus filiformis (bacteria) / Gene: THFILI_06775 / Plasmid: pET-28c+ / Production host:  |
|---|---|
| #2: Chemical | ChemComp-ZN / |
| #3: Chemical | ChemComp-B12 / |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: 2D ARRAY / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cobalamin-dependent methionine synthase holoprotein / Type: COMPLEX Details: Cobalamin-dependent methionine synthase (MetH) in complex with B12 and Zinc Entity ID: #1 / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.132539 MDa / Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:   Thermus filiformis (bacteria) Thermus filiformis (bacteria) | ||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||
| Buffer solution | pH: 7.6 / Details: 50 mM HEPES, 150 mM NaCl, 2.5 mM DTT pH 7.6 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 0.264 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES Details: Sample was mixed with equimolar commercial horse spleen apoferritin for freezing. | ||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE-PROPANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 81000 X / Nominal defocus max: 2500 nm / Nominal defocus min: 800 nm / Cs: 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 2.5 sec. / Electron dose: 65 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) / Num. of grids imaged: 2 / Num. of real images: 12768 Details: Images were collected in movie mode with a total of 50 frames over 2.5 seconds. |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 30 eV |
| Image scans | Width: 5760 / Height: 5092 |
- Processing
Processing
| EM software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: cryoSPARC patch CTF / Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 9994870 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 257706 / Num. of class averages: 1 / Symmetry type: POINT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL Details: Initial model used was the 3 N-terminal domains of AlphaFold database model A0A1Q9SZ17. Each domain was fit individually by rigid-body fitting in Chimera. The cobalamin ligand was initially ...Details: Initial model used was the 3 N-terminal domains of AlphaFold database model A0A1Q9SZ17. Each domain was fit individually by rigid-body fitting in Chimera. The cobalamin ligand was initially fit by alignment of the 1BMT crystal structure to the appropriate binding region. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Accession code: A0A1Q9SZ17 / Chain residue range: 2-896 / Source name: AlphaFold / Type: in silico model |
 Movie
Movie Controller
Controller


 PDBj
PDBj