[English] 日本語
 Yorodumi
Yorodumi- PDB-7zk3: Structure of 1PBC- and calcium-bound mTMEM16A(ac) chloride channe... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7zk3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of 1PBC- and calcium-bound mTMEM16A(ac) chloride channel at 2.85 A resolution | |||||||||
 Components Components | Anoctamin-1 | |||||||||
 Keywords Keywords | MEMBRANE PROTEIN / calcium-activated chloride channel / anoctamin-1 | |||||||||
| Function / homology |  Function and homology information Function and homology informationglial cell projection elongation / trachea development / mucus secretion / intracellularly calcium-gated chloride channel activity / voltage-gated chloride channel activity / Stimuli-sensing channels / chloride transport / chloride channel activity / detection of temperature stimulus involved in sensory perception of pain / positive regulation of insulin secretion involved in cellular response to glucose stimulus ...glial cell projection elongation / trachea development / mucus secretion / intracellularly calcium-gated chloride channel activity / voltage-gated chloride channel activity / Stimuli-sensing channels / chloride transport / chloride channel activity / detection of temperature stimulus involved in sensory perception of pain / positive regulation of insulin secretion involved in cellular response to glucose stimulus / chloride channel complex / chloride transmembrane transport / regulation of membrane potential / cell projection / establishment of localization in cell / presynaptic membrane / cellular response to heat / phospholipase C-activating G protein-coupled receptor signaling pathway / apical plasma membrane / external side of plasma membrane / glutamatergic synapse / protein homodimerization activity / metal ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.85 Å | |||||||||
 Authors Authors | Lam, A.K.M. / Rutz, S. / Dutzler, R. | |||||||||
| Funding support | European Union,  Switzerland, 2items Switzerland, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Inhibition mechanism of the chloride channel TMEM16A by the pore blocker 1PBC. Authors: Andy K M Lam / Sonja Rutz / Raimund Dutzler /  Abstract: TMEM16A, a calcium-activated chloride channel involved in multiple cellular processes, is a proposed target for diseases such as hypertension, asthma, and cystic fibrosis. Despite these therapeutic ...TMEM16A, a calcium-activated chloride channel involved in multiple cellular processes, is a proposed target for diseases such as hypertension, asthma, and cystic fibrosis. Despite these therapeutic promises, its pharmacology remains poorly understood. Here, we present a cryo-EM structure of TMEM16A in complex with the channel blocker 1PBC and a detailed functional analysis of its inhibition mechanism. A pocket located external to the neck region of the hourglass-shaped pore is responsible for open-channel block by 1PBC and presumably also by its structural analogs. The binding of the blocker stabilizes an open-like conformation of the channel that involves a rearrangement of several pore helices. The expansion of the outer pore enhances blocker sensitivity and enables 1PBC to bind at a site within the transmembrane electric field. Our results define the mechanism of inhibition and gating and will facilitate the design of new, potent TMEM16A modulators. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7zk3.cif.gz 7zk3.cif.gz | 283.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7zk3.ent.gz pdb7zk3.ent.gz | 218.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7zk3.json.gz 7zk3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7zk3_validation.pdf.gz 7zk3_validation.pdf.gz | 1.1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7zk3_full_validation.pdf.gz 7zk3_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  7zk3_validation.xml.gz 7zk3_validation.xml.gz | 55 KB | Display | |
| Data in CIF |  7zk3_validation.cif.gz 7zk3_validation.cif.gz | 78.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zk/7zk3 https://data.pdbj.org/pub/pdb/validation_reports/zk/7zk3 ftp://data.pdbj.org/pub/pdb/validation_reports/zk/7zk3 ftp://data.pdbj.org/pub/pdb/validation_reports/zk/7zk3 | HTTPS FTP |
-Related structure data
| Related structure data |  14753MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
NCS oper: (Code: givenMatrix: (-0.999980068895, -0.0038673588816, -0.00499052590843), (0.0038441086586, -0.999981753054, 0.004660084584), (-0.00500845706608, 0.00464080757951, 0.99997668886)Vector: ...NCS oper: (Code: given Matrix: (-0.999980068895, -0.0038673588816, -0.00499052590843), Vector: |
- Components
Components
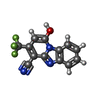
| #1: Protein | Mass: 111058.992 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: Q8BHY3 Homo sapiens (human) / References: UniProt: Q8BHY3#2: Chemical | ChemComp-CA / #3: Chemical | Has ligand of interest | Y | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: mouse TMEM16A ac splice variant in complex with 1PBC and calcium Type: ORGANELLE OR CELLULAR COMPONENT / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  |
| Source (recombinant) | Organism:  Homo sapiens (human) / Cell: GnTI- HEK293S Homo sapiens (human) / Cell: GnTI- HEK293S |
| Buffer solution | pH: 7.4 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid type: Quantifoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE-PROPANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 130000 X / Nominal defocus max: 2400 nm / Nominal defocus min: 1000 nm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 61 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.85 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 101813 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||
| Displacement parameters | Biso mean: 36.56 Å2 | ||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||
| Refine LS restraints NCS | Type: NCS constraints / Rms dev position: 0.000706321995647 Å |
 Movie
Movie Controller
Controller


 PDBj
PDBj