+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7pi5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Unstacked stretched Dunaliella PSII | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | PHOTOSYNTHESIS / green algae / photosystem II / thylakoid / oxygen evolving complex / cryo-EM / stacking | |||||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthesis, light harvesting / oxygen evolving activity / photosystem II stabilization / photosystem II reaction center / photosystem II / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosynthetic electron transport chain / photosystem I / photosystem II / response to herbicide ...photosynthesis, light harvesting / oxygen evolving activity / photosystem II stabilization / photosystem II reaction center / photosystem II / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosynthetic electron transport chain / photosystem I / photosystem II / response to herbicide / chlorophyll binding / photosynthetic electron transport in photosystem II / phosphate ion binding / photosynthesis, light reaction / chloroplast thylakoid membrane / photosynthesis / electron transfer activity / protein stabilization / iron ion binding / heme binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Dunaliella salina (plant) Dunaliella salina (plant) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.78 Å | |||||||||
 Authors Authors | Caspy, I. / Fadeeva, M. / Mazor, Y. / Nelson, N. | |||||||||
| Funding support |  Israel, 2items Israel, 2items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Structure of photosystem II reveals conformational flexibility of stacked and unstacked supercomplexes. Authors: Ido Caspy / Maria Fadeeva / Yuval Mazor / Nathan Nelson /   Abstract: Photosystem II (PSII) generates an oxidant whose redox potential is high enough to enable water oxidation , a substrate so abundant that it assures a practically unlimited electron source for life on ...Photosystem II (PSII) generates an oxidant whose redox potential is high enough to enable water oxidation , a substrate so abundant that it assures a practically unlimited electron source for life on earth . Our knowledge on the mechanism of water photooxidation was greatly advanced by high-resolution structures of prokaryotic PSII . Here, we show high-resolution cryogenic electron microscopy (cryo-EM) structures of eukaryotic PSII from the green alga at two distinct conformations. The conformers are also present in stacked PSII, exhibiting flexibility that may be relevant to the grana formation in chloroplasts of the green lineage. CP29, one of PSII associated light-harvesting antennae, plays a major role in distinguishing the two conformations of the supercomplex. We also show that the stacked PSII dimer, a form suggested to support the organisation of thylakoid membranes , can appear in many different orientations providing a flexible stacking mechanism for the arrangement of grana stacks in thylakoids. Our findings provide a structural basis for the heterogenous nature of the eukaryotic PSII on multiple levels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7pi5.cif.gz 7pi5.cif.gz | 1.6 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7pi5.ent.gz pdb7pi5.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7pi5.json.gz 7pi5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pi/7pi5 https://data.pdbj.org/pub/pdb/validation_reports/pi/7pi5 ftp://data.pdbj.org/pub/pdb/validation_reports/pi/7pi5 ftp://data.pdbj.org/pub/pdb/validation_reports/pi/7pi5 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  13430MC  7pi0C  7pinC  7piwC  7pnkC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Photosystem II ... , 13 types, 26 molecules AaBbVvCcDdHhIiJjKkLlMmTtZz
| #1: Protein | Mass: 37291.488 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FY08, photosystem II Dunaliella salina (plant) / References: UniProt: D0FY08, photosystem II#2: Protein | Mass: 53289.516 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FY05 Dunaliella salina (plant) / References: UniProt: D0FY05#3: Protein/peptide | Mass: 3217.949 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FY13 Dunaliella salina (plant) / References: UniProt: D0FY13#4: Protein | Mass: 49128.992 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FXY3 Dunaliella salina (plant) / References: UniProt: D0FXY3#5: Protein | Mass: 38965.383 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: A0A1C8XRK9, photosystem II Dunaliella salina (plant) / References: UniProt: A0A1C8XRK9, photosystem II#8: Protein | Mass: 7124.381 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: A0A1C8XRP3 Dunaliella salina (plant) / References: UniProt: A0A1C8XRP3#9: Protein/peptide | Mass: 3929.648 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FXX5 Dunaliella salina (plant) / References: UniProt: D0FXX5#10: Protein/peptide | Mass: 3737.500 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: A0A1C8XRM8 Dunaliella salina (plant) / References: UniProt: A0A1C8XRM8#11: Protein/peptide | Mass: 4159.991 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FXX2 Dunaliella salina (plant) / References: UniProt: D0FXX2#12: Protein/peptide | Mass: 4417.203 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FY19 Dunaliella salina (plant) / References: UniProt: D0FY19#13: Protein/peptide | Mass: 3437.093 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FXZ3 Dunaliella salina (plant) / References: UniProt: D0FXZ3#16: Protein/peptide | Mass: 3478.194 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FY04 Dunaliella salina (plant) / References: UniProt: D0FY04#19: Protein | Mass: 6430.616 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FXZ2 Dunaliella salina (plant) / References: UniProt: D0FXZ2 |
|---|
-Cytochrome b559 subunit ... , 2 types, 4 molecules EeFf
| #6: Protein | Mass: 8743.847 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: D0FY01 Dunaliella salina (plant) / References: UniProt: D0FY01#7: Protein/peptide | Mass: 3549.299 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: A0A1C8XRP4 Dunaliella salina (plant) / References: UniProt: A0A1C8XRP4 |
|---|
-Protein , 7 types, 14 molecules OoPpNnGgRrSsYy
| #14: Protein | Mass: 25874.908 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) Dunaliella salina (plant)#15: Protein | Mass: 20425.707 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) Dunaliella salina (plant)#20: Protein | Mass: 24075.082 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) Dunaliella salina (plant)#21: Protein | Mass: 23719.543 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: A1XKU7 Dunaliella salina (plant) / References: UniProt: A1XKU7#22: Protein | Mass: 21748.461 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) Dunaliella salina (plant)#23: Protein | Mass: 26233.607 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: A0A7S3VRZ8 Dunaliella salina (plant) / References: UniProt: A0A7S3VRZ8#24: Protein | Mass: 23537.566 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: A0A6S8N9J6 Dunaliella salina (plant) / References: UniProt: A0A6S8N9J6 |
|---|
-Protein/peptide , 3 types, 6 molecules WwXxUu
| #17: Protein/peptide | Mass: 4698.315 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: A0A7S3QU88 Dunaliella salina (plant) / References: UniProt: A0A7S3QU88#18: Protein/peptide | Mass: 2854.407 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) / References: UniProt: A0A7S3VKF3 Dunaliella salina (plant) / References: UniProt: A0A7S3VKF3#25: Protein/peptide | Mass: 3219.625 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Dunaliella salina (plant) Dunaliella salina (plant) |
|---|
-Sugars , 1 types, 8 molecules 
| #38: Sugar | ChemComp-DGD / |
|---|
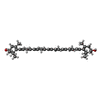
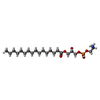
+Non-polymers , 26 types, 1053 molecules 
















































 Movie
Movie Controller
Controller







 PDBj
PDBj
































