+ Open data
Open data
- Basic information
Basic information
| Entry | 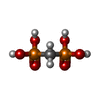 Database: PDB chemical components / ID: MDN Database: PDB chemical components / ID: MDN |
|---|---|
| Name | Name: |
-Chemical information
| Composition |  | ||||
|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAIN / Three letter code: MDN / Model coordinates PDB-ID: 1HUZ | ||||
| History |
| ||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | | OpenEye OEToolkits 1.5.0 | |
|---|
-PDB entries
Showing all 6 items

PDB-1huz: 
CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP

PDB-3hxy: 
Crystal Structure of catalytic fragment of E. coli AlaRS in complex with AMPPCP, Ala-AMP and PCP

PDB-5tlh: 
Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with the inhibitor 2-naphthol 6-bisphosphonate
PDB-5ysp: 
Pyrophosphate-dependent kinase in the ribokinase family complexed with a pyrophosphate analog and myo-inositol

PDB-6s83: 
Crystal structure of methionine adenosyltransferase from Pyrococcus furiosus in complex with AMPPCP, SAM, and PCP

PDB-7e4l: 
Conversion of pyrophosphate-dependent myo-inositol-1 kinase into myo-inositol-3 kinase by N78L/S89L mutation
 Movie
Movie Controller
Controller



