[English] 日本語
 Yorodumi
Yorodumi- EMDB-9131: Cryo-EM structure of the ZIKV virion in complex with Fab fragment... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9131 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the ZIKV virion in complex with Fab fragments of the potently neutralizing human monoclonal antibody ZIKV-195 | |||||||||
 Map data Map data | ZIKV virion in complex with Fab fragments of the potently neutralizing human monoclonal antibody ZIKV-195. Different contour levels are required to visualize the viral components and the bound Fab molecules. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus / monoclonal antibody / complex / VIRUS-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationflavivirin / immunoglobulin complex / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / negative regulation of innate immune response / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / 4 iron, 4 sulfur cluster binding ...flavivirin / immunoglobulin complex / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / negative regulation of innate immune response / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / 4 iron, 4 sulfur cluster binding / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / clathrin-dependent endocytosis of virus by host cell / mRNA (nucleoside-2'-O-)-methyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / adaptive immune response / molecular adaptor activity / RNA helicase activity / host cell perinuclear region of cytoplasm / protein dimerization activity / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / immune response / induction by virus of host autophagy / RNA-directed RNA polymerase / viral RNA genome replication / serine-type endopeptidase activity / RNA-dependent RNA polymerase activity / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / centrosome / lipid binding / viral envelope / host cell nucleus / virion attachment to host cell / GTP binding / structural molecule activity / virion membrane / ATP hydrolysis activity / proteolysis / extracellular space / extracellular region / ATP binding / membrane / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013) Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Long F / Rossmann MG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Structural basis of a potent human monoclonal antibody against Zika virus targeting a quaternary epitope. Authors: Feng Long / Michael Doyle / Estefania Fernandez / Andrew S Miller / Thomas Klose / Madhumati Sevvana / Aubrey Bryan / Edgar Davidson / Benjamin J Doranz / Richard J Kuhn / Michael S Diamond ...Authors: Feng Long / Michael Doyle / Estefania Fernandez / Andrew S Miller / Thomas Klose / Madhumati Sevvana / Aubrey Bryan / Edgar Davidson / Benjamin J Doranz / Richard J Kuhn / Michael S Diamond / James E Crowe / Michael G Rossmann /  Abstract: Zika virus (ZIKV) is a major human pathogen and member of the genus in the Flaviviridae family. In contrast to most other insect-transmitted flaviviruses, ZIKV also can be transmitted sexually and ...Zika virus (ZIKV) is a major human pathogen and member of the genus in the Flaviviridae family. In contrast to most other insect-transmitted flaviviruses, ZIKV also can be transmitted sexually and from mother to fetus in humans. During recent outbreaks, ZIKV infections have been linked to microcephaly, congenital disease, and Guillain-Barré syndrome. Neutralizing antibodies have potential as therapeutic agents. We report here a 4-Å-resolution cryo-electron microscopy structure of the ZIKV virion in complex with Fab fragments of the potently neutralizing human monoclonal antibody ZIKV-195. The footprint of the ZIKV-195 Fab fragment expands across two adjacent envelope (E) protein protomers. ZIKV neutralization by this antibody is presumably accomplished by cross-linking the E proteins, which likely prevents formation of E protein trimers required for fusion of the viral and cellular membranes. A single dose of ZIKV-195 administered 5 days after virus inoculation showed marked protection against lethality in a stringent mouse model of infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9131.map.gz emd_9131.map.gz | 478.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9131-v30.xml emd-9131-v30.xml emd-9131.xml emd-9131.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
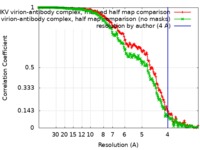
| FSC (resolution estimation) |  emd_9131_fsc_1.xml emd_9131_fsc_1.xml emd_9131_fsc_2.xml emd_9131_fsc_2.xml | 21 KB 21 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_9131.png emd_9131.png | 287.6 KB | ||
| Filedesc metadata |  emd-9131.cif.gz emd-9131.cif.gz | 7.1 KB | ||
| Others |  emd_9131_half_map_1.map.gz emd_9131_half_map_1.map.gz emd_9131_half_map_2.map.gz emd_9131_half_map_2.map.gz | 130.1 MB 130.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9131 http://ftp.pdbj.org/pub/emdb/structures/EMD-9131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9131 | HTTPS FTP |
-Validation report
| Summary document |  emd_9131_validation.pdf.gz emd_9131_validation.pdf.gz | 980 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9131_full_validation.pdf.gz emd_9131_full_validation.pdf.gz | 979.6 KB | Display | |
| Data in XML |  emd_9131_validation.xml.gz emd_9131_validation.xml.gz | 19.7 KB | Display | |
| Data in CIF |  emd_9131_validation.cif.gz emd_9131_validation.cif.gz | 23.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9131 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9131 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9131 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9131 | HTTPS FTP |
-Related structure data
| Related structure data |  6midMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9131.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9131.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ZIKV virion in complex with Fab fragments of the potently neutralizing human monoclonal antibody ZIKV-195. Different contour levels are required to visualize the viral components and the bound Fab molecules. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
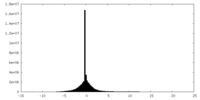
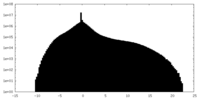
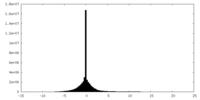
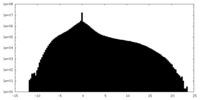
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map #1
| File | emd_9131_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map #2
| File | emd_9131_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ZIKV virion in complex with Fab fragments of the potently neutral...
| Entire | Name: ZIKV virion in complex with Fab fragments of the potently neutralizing human monoclonal antibody ZIKV-195 |
|---|---|
| Components |
|
-Supramolecule #1: ZIKV virion in complex with Fab fragments of the potently neutral...
| Supramolecule | Name: ZIKV virion in complex with Fab fragments of the potently neutralizing human monoclonal antibody ZIKV-195 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Purified from infected Vero cells overexpressing the furin protease |
|---|
-Supramolecule #3: monoclonal antibody ZIKV-195
| Supramolecule | Name: monoclonal antibody ZIKV-195 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013)
| Supramolecule | Name: Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013) type: virus / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 / NCBI-ID: 2043570 Sci species name: Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013) Sci species strain: isolate ZIKV/Human/French Polynesia/10087PF/2013 Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: E protein
| Macromolecule | Name: E protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013) Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013)Strain: isolate ZIKV/Human/French Polynesia/10087PF/2013 |
| Molecular weight | Theoretical: 54.444051 KDa |
| Sequence | String: IRCIGVSNRD FVEGMSGGTW VDVVLEHGGC VTVMAQDKPT VDIELVTTTV SNMAEVRSYC YEASISDMAS DSRCPTQGEA YLDKQSDTQ YVCKRTLVDR GWGNGCGLFG KGSLVTCAKF ACSKKMTGKS IQPENLEYRI MLSVHGSQHS GMIVNDTGHE T DENRAKVE ...String: IRCIGVSNRD FVEGMSGGTW VDVVLEHGGC VTVMAQDKPT VDIELVTTTV SNMAEVRSYC YEASISDMAS DSRCPTQGEA YLDKQSDTQ YVCKRTLVDR GWGNGCGLFG KGSLVTCAKF ACSKKMTGKS IQPENLEYRI MLSVHGSQHS GMIVNDTGHE T DENRAKVE ITPNSPRAEA TLGGFGSLGL DCEPRTGLDF SDLYYLTMNN KHWLVHKEWF HDIPLPWHAG ADTGTPHWNN KE ALVEFKD AHAKRQTVVV LGSQEGAVHT ALAGALEAEM DGAKGRLSSG HLKCRLKMDK LRLKGVSYSL CTAAFTFTKI PAE TLHGTV TVEVQYAGTD GPCKVPAQMA VDMQTLTPVG RLITANPVIT ESTENSKMML ELDPPFGDSY IVIGVGEKKI THHW HRSGS TIGKAFEATV RGAKRMAVLG DTAWDFGSVG GALNSLGKGI HQIFGAAFKS LFGGMSWFSQ ILIGTLLMWL GLNTK NGSI SLMCLALGGV LIFLSTAVSA UniProtKB: Genome polyprotein |
-Macromolecule #2: M protein
| Macromolecule | Name: M protein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013) Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013)Strain: isolate ZIKV/Human/French Polynesia/10087PF/2013 |
| Molecular weight | Theoretical: 8.496883 KDa |
| Sequence | String: AVTLPSHSTR KLQTRSQTWL ESREYTKHLI RVENWIFRNP GFALAAAAIA WLLGSSTSQK VIYLVMILLI APAYS UniProtKB: Genome polyprotein |
-Macromolecule #3: monoclonal antibody ZIKV-195 heavy chain
| Macromolecule | Name: monoclonal antibody ZIKV-195 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.148739 KDa |
| Sequence | String: QVQLVESGGG VVHPGRSLRL SCAASGFTFS SSAMHWVRQA PGKGLEWVAV ISYDGSNKYY GDSVKGRFTI SRDNSKNTLY LQMHSLRAE DTAVYYCAKD RDAYNTVGYF AYYYGMDVWG QGTLVTVSS |
-Macromolecule #4: monoclonal antibody ZIKV-195 light chain
| Macromolecule | Name: monoclonal antibody ZIKV-195 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.750902 KDa |
| Sequence | String: QSVLTQPPSV SEAPRQRVTI SCSGSSSNIG NNAVNWYQQL PGKAPKLLIY YDDLLPSGVS DRFSGSKSGT SASLAISGLQ SEDEADYYC AAWDDSLTRY VFGTGTKVTV L UniProtKB: Immunoglobulin lambda variable 1-36 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.00 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: NTE buffer at pH 8.0 | ||||||||||||
| Grid | Details: unspecified | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 | ||||||||||||
| Details | Fab fragments were mixed with purified mature virus particles to make complexes. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 1691 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | The occupancy of bound Fabs was low. Due to poor density, only the variable domain of Fab molecule was modeled and fit into one of the three Fab binding sites on each icosahedral asymmetric unit. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-6mid: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)