[English] 日本語
 Yorodumi
Yorodumi- EMDB-8924: Structure of the Salmonella SPI-1 type III secretion injectisome ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8924 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Salmonella SPI-1 type III secretion injectisome needle filament | |||||||||
 Map data Map data | Cryo-EM reconstruction of the Salmonella SPI-1 type III secretion injectisome needle filament | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type III secretion system / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtype III protein secretion system complex / protein secretion by the type III secretion system / cell surface / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Hu J / Hong C | |||||||||
| Funding support |  Canada, Canada,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Authors: J Hu / L J Worrall / C Hong / M Vuckovic / C E Atkinson / N Caveney / Z Yu / N C J Strynadka /   Abstract: The bacterial type III secretion system, or injectisome, is a syringe shaped nanomachine essential for the virulence of many disease causing Gram-negative bacteria. At the core of the injectisome ...The bacterial type III secretion system, or injectisome, is a syringe shaped nanomachine essential for the virulence of many disease causing Gram-negative bacteria. At the core of the injectisome structure is the needle complex, a continuous channel formed by the highly oligomerized inner and outer membrane hollow rings and a polymerized helical needle filament which spans through and projects into the infected host cell. Here we present the near-atomic resolution structure of a needle complex from the prototypical Salmonella Typhimurium SPI-1 type III secretion system, with local masking protocols allowing for model building and refinement of the major membrane spanning components of the needle complex base in addition to an isolated needle filament. This work provides significant insight into injectisome structure and assembly and importantly captures the molecular basis for substrate induced gating in the giant outer membrane secretin portal family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8924.map.gz emd_8924.map.gz | 2.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8924-v30.xml emd-8924-v30.xml emd-8924.xml emd-8924.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8924.png emd_8924.png | 42 KB | ||
| Filedesc metadata |  emd-8924.cif.gz emd-8924.cif.gz | 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8924 http://ftp.pdbj.org/pub/emdb/structures/EMD-8924 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8924 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8924 | HTTPS FTP |
-Related structure data
| Related structure data |  6dwbMC  8913C  8914C  8915C  6duzC  6dv3C  6dv6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8924.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8924.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the Salmonella SPI-1 type III secretion injectisome needle filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
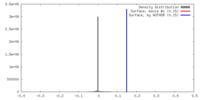
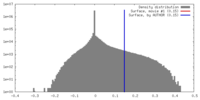
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PrgI helical filament
| Entire | Name: PrgI helical filament |
|---|---|
| Components |
|
-Supramolecule #1: PrgI helical filament
| Supramolecule | Name: PrgI helical filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) |
-Macromolecule #1: Protein PrgI
| Macromolecule | Name: Protein PrgI / type: protein_or_peptide / ID: 1 / Number of copies: 30 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) |
| Molecular weight | Theoretical: 8.864868 KDa |
| Sequence | String: MATPWSGYLD DVSAKFDTGV DNLQTQVTEA LDKLAAKPSD PALLAAYQSK LSEYNLYRNA QSNTVKVFKD IDAAIIQNFR UniProtKB: SPI-1 type 3 secretion system needle filament protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Cs corrector was used. / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 3753 / Average exposure time: 0.25 sec. / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.6 µm / Calibrated defocus min: 1.1 µm / Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6dwb: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)