+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8745 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. cerevisiae Hsp104:casein complex, Middle Domain Conformation | |||||||||
 Map data Map data | Hsp104:casein complex, Middle Domain Conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hsp104 / cryoem / AAA+ / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationtrehalose metabolic process / TRC complex / protein folding in endoplasmic reticulum / cellular heat acclimation / post-translational protein targeting to endoplasmic reticulum membrane / stress granule disassembly / : / protein unfolding / nuclear periphery / ADP binding ...trehalose metabolic process / TRC complex / protein folding in endoplasmic reticulum / cellular heat acclimation / post-translational protein targeting to endoplasmic reticulum membrane / stress granule disassembly / : / protein unfolding / nuclear periphery / ADP binding / unfolded protein binding / protein-folding chaperone binding / cellular response to heat / protein refolding / ATP hydrolysis activity / ATP binding / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.7 Å | |||||||||
 Authors Authors | Gates SN / Yokom AL | |||||||||
 Citation Citation |  Journal: Science / Year: 2017 Journal: Science / Year: 2017Title: Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Authors: Stephanie N Gates / Adam L Yokom / JiaBei Lin / Meredith E Jackrel / Alexandrea N Rizo / Nathan M Kendsersky / Courtney E Buell / Elizabeth A Sweeny / Korrie L Mack / Edward Chuang / Mariana ...Authors: Stephanie N Gates / Adam L Yokom / JiaBei Lin / Meredith E Jackrel / Alexandrea N Rizo / Nathan M Kendsersky / Courtney E Buell / Elizabeth A Sweeny / Korrie L Mack / Edward Chuang / Mariana P Torrente / Min Su / James Shorter / Daniel R Southworth /  Abstract: Hsp100 polypeptide translocases are conserved members of the AAA+ family (adenosine triphosphatases associated with diverse cellular activities) that maintain proteostasis by unfolding aberrant and ...Hsp100 polypeptide translocases are conserved members of the AAA+ family (adenosine triphosphatases associated with diverse cellular activities) that maintain proteostasis by unfolding aberrant and toxic proteins for refolding or proteolytic degradation. The Hsp104 disaggregase from solubilizes stress-induced amorphous aggregates and amyloids. The structural basis for substrate recognition and translocation is unknown. Using a model substrate (casein), we report cryo-electron microscopy structures at near-atomic resolution of Hsp104 in different translocation states. Substrate interactions are mediated by conserved, pore-loop tyrosines that contact an 80-angstrom-long unfolded polypeptide along the axial channel. Two protomers undergo a ratchet-like conformational change that advances pore loop-substrate interactions by two amino acids. These changes are coupled to activation of specific nucleotide hydrolysis sites and, when transmitted around the hexamer, reveal a processive rotary translocation mechanism and substrate-responsive flexibility during Hsp104-catalyzed disaggregation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8745.map.gz emd_8745.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8745-v30.xml emd-8745-v30.xml emd-8745.xml emd-8745.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8745.png emd_8745.png | 65 KB | ||
| Filedesc metadata |  emd-8745.cif.gz emd-8745.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8745 http://ftp.pdbj.org/pub/emdb/structures/EMD-8745 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8745 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8745 | HTTPS FTP |
-Validation report
| Summary document |  emd_8745_validation.pdf.gz emd_8745_validation.pdf.gz | 392.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8745_full_validation.pdf.gz emd_8745_full_validation.pdf.gz | 392.4 KB | Display | |
| Data in XML |  emd_8745_validation.xml.gz emd_8745_validation.xml.gz | 5.3 KB | Display | |
| Data in CIF |  emd_8745_validation.cif.gz emd_8745_validation.cif.gz | 6.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8745 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8745 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8745 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8745 | HTTPS FTP |
-Related structure data
| Related structure data |  5vy9MC  8697C  8744C  8746C  5vjhC  5vy8C  5vyaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8745.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8745.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hsp104:casein complex, Middle Domain Conformation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
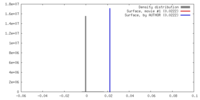
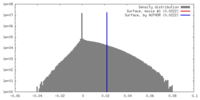
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : S. cerevisiae Hsp104:casein complex, Middle Domain Conformation
| Entire | Name: S. cerevisiae Hsp104:casein complex, Middle Domain Conformation |
|---|---|
| Components |
|
-Supramolecule #1: S. cerevisiae Hsp104:casein complex, Middle Domain Conformation
| Supramolecule | Name: S. cerevisiae Hsp104:casein complex, Middle Domain Conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
-Supramolecule #2: Hsp104
| Supramolecule | Name: Hsp104 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|
-Supramolecule #3: casein
| Supramolecule | Name: casein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Heat shock protein 104
| Macromolecule | Name: Heat shock protein 104 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 102.173961 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNDQTQFTER ALTILTLAQK LASDHQHPQL QPIHILAAFI ETPEDGSVPY LQNLIEKGRY DYDLFKKVVN RNLVRIPQQQ PAPAEITPS YALGKVLQDA AKIQKQQKDS FIAQDHILFA LFNDSSIQQI FKEAQVDIEA IKQQALELRG NTRIDSRGAD T NTPLEYLS ...String: MNDQTQFTER ALTILTLAQK LASDHQHPQL QPIHILAAFI ETPEDGSVPY LQNLIEKGRY DYDLFKKVVN RNLVRIPQQQ PAPAEITPS YALGKVLQDA AKIQKQQKDS FIAQDHILFA LFNDSSIQQI FKEAQVDIEA IKQQALELRG NTRIDSRGAD T NTPLEYLS KYAIDMTEQA RQGKLDPVIG REEEIRSTIR VLARRIKSNP CLIGEPGIGK TAIIEGVAQR IIDDDVPTIL QG AKLFSLD LAALTAGAKY KGDFEERFKG VLKEIEESKT LIVLFIDEIH MLMGNGKDDA ANILKPALSR GQLKVIGATT NNE YRSIVE KDGAFERRFQ KIEVAEPSVR QTVAILRGLQ PKYEIHHGVR ILDSALVTAA QLAKRYLPYR RLPDSALDLV DISC AGVAV ARDSKPEELD SKERQLQLIQ VEIKALERDE DADSTTKDRL KLARQKEASL QEELEPLRQR YNEEKHGHEE LTQAK KKLD ELENKALDAE RRYDTATAAD LRYFAIPDIK KQIEKLEDQV AEEERRAGAN SMIQNVVDSD TISETAARLT GIPVKK LSE SENEKLIHME RDLSSEVVGQ MDAIKAVSNA VRLSRSGLAN PRQPASFLFL GLSGSGKTEL AKKVAGFLFN DEDMMIR VD CSELSEKYAV SKLLGTTAGY VGYDEGGFLT NQLQYKPYSV LLFDEVEKAH PDVLTVMLQM LDDGRITSGQ GKTIDCSN C IVIMTSNLGA EFINSQQGSK IQESTKNLVM GAVRQHFRPE FLNRISSIVI FNKLSRKAIH KIVDIRLKEI EERFEQNDK HYKLNLTQEA KDFLAKYGYS DDMGARPLNR LIQNEILNKL ALRILKNEIK DKETVNVVLK KGKSRDENVP EEAEECLEVL PNHEATIGA DTLGDDDNED SMEIDDDLD UniProtKB: Heat shock protein 104 |
-Macromolecule #2: Alpha-S1-casein
| Macromolecule | Name: Alpha-S1-casein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.400951 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 12 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 0.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)