+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8405 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The archaeal flagellum of Methanospirillum hungatei strain JF1. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cell motility and adhesion / CELL ADHESION | |||||||||
| Function / homology | archaeal-type flagellum / Flagellin, archaea / Archaeal-type flagellin / Flagellin/pilin, N-terminal / archaeal or bacterial-type flagellum-dependent cell motility / structural molecule activity / membrane / Flagellin Function and homology information Function and homology information | |||||||||
| Biological species |  Methanospirillum hungatei JF-1 (archaea) / Methanospirillum hungatei JF-1 (archaea) /  Methanospirillum hungatei JF-1 (strain ATCC 27890 / DSM 864 / NBRC 100397 / JF-1) (archaea) Methanospirillum hungatei JF-1 (strain ATCC 27890 / DSM 864 / NBRC 100397 / JF-1) (archaea) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Poweleit N / Peng G | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2016 Journal: Nat Microbiol / Year: 2016Title: CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Authors: Nicole Poweleit / Peng Ge / Hong H Nguyen / Rachel R Ogorzalek Loo / Robert P Gunsalus / Z Hong Zhou /  Abstract: Archaea use flagella known as archaella-distinct both in protein composition and structure from bacterial flagella-to drive cell motility, but the structural basis of this function is unknown. Here, ...Archaea use flagella known as archaella-distinct both in protein composition and structure from bacterial flagella-to drive cell motility, but the structural basis of this function is unknown. Here, we report an atomic model of the archaella, based on the cryo electron microscopy (cryoEM) structure of the Methanospirillum hungatei archaellum at 3.4 Å resolution. Each archaellum contains ∼61,500 archaellin subunits organized into a curved helix with a diameter of 10 nm and average length of 10,000 nm. The tadpole-shaped archaellin monomer has two domains, a β-barrel domain and a long, mildly kinked α-helix tail. Our structure reveals multiple post-translational modifications to the archaella, including six O-linked glycans and an unusual N-linked modification. The extensive interactions among neighbouring archaellins explain how the long but thin archaellum maintains the structural integrity required for motility-driving rotation. These extensive inter-subunit interactions and the absence of a central pore in the archaellum distinguish it from both the bacterial flagellum and type IV pili. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8405.map.gz emd_8405.map.gz | 57.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8405-v30.xml emd-8405-v30.xml emd-8405.xml emd-8405.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8405.png emd_8405.png | 147.9 KB | ||
| Filedesc metadata |  emd-8405.cif.gz emd-8405.cif.gz | 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8405 http://ftp.pdbj.org/pub/emdb/structures/EMD-8405 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8405 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8405 | HTTPS FTP |
-Related structure data
| Related structure data |  5tfyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8405.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8405.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
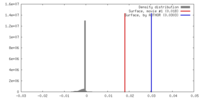
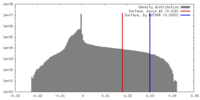
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The flagellum of the archaea Methanospirillum hungatei
| Entire | Name: The flagellum of the archaea Methanospirillum hungatei |
|---|---|
| Components |
|
-Supramolecule #1: The flagellum of the archaea Methanospirillum hungatei
| Supramolecule | Name: The flagellum of the archaea Methanospirillum hungatei type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Methanospirillum hungatei JF-1 (archaea) / Strain: JF1 Methanospirillum hungatei JF-1 (archaea) / Strain: JF1 |
| Molecular weight | Theoretical: 20.048 kDa/nm |
-Macromolecule #1: Flagellin
| Macromolecule | Name: Flagellin / type: protein_or_peptide / ID: 1 / Number of copies: 26 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Methanospirillum hungatei JF-1 (strain ATCC 27890 / DSM 864 / NBRC 100397 / JF-1) (archaea) Methanospirillum hungatei JF-1 (strain ATCC 27890 / DSM 864 / NBRC 100397 / JF-1) (archaea)Strain: ATCC 27890 / DSM 864 / NBRC 100397 / JF-1 |
| Molecular weight | Theoretical: 17.524129 KDa |
| Sequence | String: FSGLEAAIVL IAFVVVAAVF SYVMLGAGFF ATQKSQEVTY SGMKQATSNL ILDGMIYGSY SKGGSGLAQL YFYVKVPEGG ETQDLKYVT YLWTKENKAV TTLTSITPTN QQLNPGARVK VTITAPTGYK PIAGQKFVLE IKPKTGASTI VTRTLSDGYN G GVII UniProtKB: Flagellin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 1 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 3-13 / Number grids imaged: 2 / Average exposure time: 0.25 sec. / Average electron dose: 2.667 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 5.2 Å Applied symmetry - Helical parameters - Δ&Phi: 108 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.33 CUT-OFF / Number images used: 90118 |
|---|---|
| Startup model | Type of model: INSILICO MODEL |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 200 |
|---|---|
| Output model |  PDB-5tfy: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)