[English] 日本語
 Yorodumi
Yorodumi- EMDB-7549: Monomer yeast ATP synthase Fo reconstituted in nanodisc generated... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7549 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Monomer yeast ATP synthase Fo reconstituted in nanodisc generated from masked refinement. | |||||||||
 Map data Map data | Monomer yeast ATP synthase Fo reconstituted in nanodisc generated from masked refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP synthase / BIOSYNTHETIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial protein degradation / mitochondrial proton-transporting ATP synthase complex assembly / proton transmembrane transporter activity / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton-transporting ATP synthase complex / proton transmembrane transport / mitochondrial intermembrane space / protein-containing complex assembly / mitochondrial inner membrane ...Mitochondrial protein degradation / mitochondrial proton-transporting ATP synthase complex assembly / proton transmembrane transporter activity / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton-transporting ATP synthase complex / proton transmembrane transport / mitochondrial intermembrane space / protein-containing complex assembly / mitochondrial inner membrane / lipid binding / mitochondrion / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
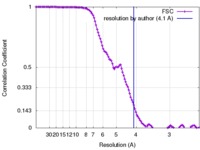
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Srivastava AP / Luo M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane. Authors: Anurag P Srivastava / Min Luo / Wenchang Zhou / Jindrich Symersky / Dongyang Bai / Melissa G Chambers / José D Faraldo-Gómez / Maofu Liao / David M Mueller /  Abstract: Mitochondrial adenosine triphosphate (ATP) synthase comprises a membrane embedded F motor that rotates to drive ATP synthesis in the F subunit. We used single-particle cryo-electron microscopy (cryo- ...Mitochondrial adenosine triphosphate (ATP) synthase comprises a membrane embedded F motor that rotates to drive ATP synthesis in the F subunit. We used single-particle cryo-electron microscopy (cryo-EM) to obtain structures of the full complex in a lipid bilayer in the absence or presence of the inhibitor oligomycin at 3.6- and 3.8-angstrom resolution, respectively. To limit conformational heterogeneity, we locked the rotor in a single conformation by fusing the F6 subunit of the stator with the δ subunit of the rotor. Assembly of the enzyme with the F6-δ fusion caused a twisting of the rotor and a 9° rotation of the F c-ring in the direction of ATP synthesis, relative to the structure of isolated F Our cryo-EM structures show how F and F are coupled, give insight into the proton translocation pathway, and show how oligomycin blocks ATP synthesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7549.map.gz emd_7549.map.gz | 101.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7549-v30.xml emd-7549-v30.xml emd-7549.xml emd-7549.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7549_fsc.xml emd_7549_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_7549.png emd_7549.png | 197.5 KB | ||
| Filedesc metadata |  emd-7549.cif.gz emd-7549.cif.gz | 5.9 KB | ||
| Others |  emd_7549_additional.map.gz emd_7549_additional.map.gz | 98.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7549 http://ftp.pdbj.org/pub/emdb/structures/EMD-7549 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7549 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7549 | HTTPS FTP |
-Related structure data
| Related structure data |  6cp7MC  7546C  7547C  7548C  6cp3C  6cp5C  6cp6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7549.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7549.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Monomer yeast ATP synthase Fo reconstituted in nanodisc generated from masked refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
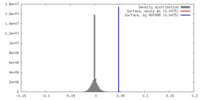
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 7549 additional.map
| File | emd_7549_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
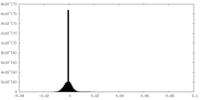
| Projections & Slices |
| ||||||||||||
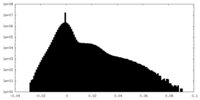
| Density Histograms |
- Sample components
Sample components
-Entire : Monomer yeast ATP synthase Fo reconstituted in nanodisc generated...
| Entire | Name: Monomer yeast ATP synthase Fo reconstituted in nanodisc generated from masked refinement. |
|---|---|
| Components |
|
-Supramolecule #1: Monomer yeast ATP synthase Fo reconstituted in nanodisc generated...
| Supramolecule | Name: Monomer yeast ATP synthase Fo reconstituted in nanodisc generated from masked refinement. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP synthase subunit 9, mitochondrial
| Macromolecule | Name: ATP synthase subunit 9, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 7.790385 KDa |
| Sequence | String: (FME)QLVLAAKYI GAGISTIGLL GAGIGIAIVF AALINGVSRN PSIKDTVFPM AILGFALSEA TGLFCLMVSF LLLFGV UniProtKB: ATP synthase subunit 9, mitochondrial |
-Macromolecule #2: ATP synthase protein 8
| Macromolecule | Name: ATP synthase protein 8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 5.825215 KDa |
| Sequence | String: MPQLVPFYFM NQLTYGFLLM ITLLILFSQF FLPMILRLYV SRLFISKL UniProtKB: ATP synthase protein 8 |
-Macromolecule #3: ATP synthase subunit a
| Macromolecule | Name: ATP synthase subunit a / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 27.90043 KDa |
| Sequence | String: SPLDQFEIRT LFGLQSSFID LSCLNLTTFS LYTIIVLLVI TSLYTLTNNN NKIIGSRWLI SQEAIYDTIM NMTKGQIGGK NWGLYFPMI FTLFMFIFIA NLISMIPYSF ALSAHLVFII SLSIVIWLGN TILGLYKHGW VFFSLFVPAG TPLPLVPLLV I IETLSYFA ...String: SPLDQFEIRT LFGLQSSFID LSCLNLTTFS LYTIIVLLVI TSLYTLTNNN NKIIGSRWLI SQEAIYDTIM NMTKGQIGGK NWGLYFPMI FTLFMFIFIA NLISMIPYSF ALSAHLVFII SLSIVIWLGN TILGLYKHGW VFFSLFVPAG TPLPLVPLLV I IETLSYFA RAISLGLRLG SNILAGHLLM VILAGLTFNF MLINLFTLVF GFVPLAMILA IMMLEFAIGI IQGYVWAILT AS YLKDAVY LH UniProtKB: ATP synthase subunit a |
-Macromolecule #4: ATP synthase subunit 4, mitochondrial
| Macromolecule | Name: ATP synthase subunit 4, mitochondrial / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 23.194498 KDa |
| Sequence | String: MSSTPEKQTD PKAKANSIIN AIPGNNILTK TGVLGTSAAA VIYAISNELY VINDESILLL TFLGFTGLVA KYLAPAYKDF ADARMKKVS DVLNASRNKH VEAVKDRIDS VSQLQNVAET TKVLFDVSKE TVELESEAFE LKQKVELAHE AKAVLDSWVR Y EASLRQLE ...String: MSSTPEKQTD PKAKANSIIN AIPGNNILTK TGVLGTSAAA VIYAISNELY VINDESILLL TFLGFTGLVA KYLAPAYKDF ADARMKKVS DVLNASRNKH VEAVKDRIDS VSQLQNVAET TKVLFDVSKE TVELESEAFE LKQKVELAHE AKAVLDSWVR Y EASLRQLE QRQLAKSVIS RVQSELGNPK FQEKVLQQSI SEIEQLLSKL K UniProtKB: ATP synthase subunit 4, mitochondrial |
-Macromolecule #5: ATP synthase subunit d, mitochondrial
| Macromolecule | Name: ATP synthase subunit d, mitochondrial / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 19.709424 KDa |
| Sequence | String: SLAKSAANKL DWAKVISSLR ITGSTATQLS SFKKRNDEAR RQLLELQSQP TEVDFSHYRS VLKNTSVIDK IESYVKQYKP VKIDASKQL QVIESFEKHA MTNAKETESL VSKELKDLQS TLDNIQSARP FDELTVDDLT KIKPEIDAKV EEMVKKGKWD V PGYKDRFG NLNVM UniProtKB: ATP synthase subunit d, mitochondrial |
-Macromolecule #6: ATP synthase subunit f, mitochondrial
| Macromolecule | Name: ATP synthase subunit f, mitochondrial / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 10.584166 KDa |
| Sequence | String: VSTLIPPKVV SSKNIGSAPN AKRIANVVHF YKSLPQGPAP AIKANTRLAR YKAKYFDGDN ASGKPLWHFA LGIIAFGYSM EYYFHLRHH KGAEEH UniProtKB: ATP synthase subunit f, mitochondrial |
-Macromolecule #7: ATP synthase subunit J, mitochondrial
| Macromolecule | Name: ATP synthase subunit J, mitochondrial / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 4.145884 KDa |
| Sequence | String: MLKRFPTPIL KVYWPFFVAG AAVYYGMSKA ADLSSNT UniProtKB: ATP synthase subunit J, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1. mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris-HCl, 150 mM NaCl, pH 8.0 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 91 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80.0 K / Max: 105.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 5935 / Average electron dose: 8.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.6 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 31000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6cp7: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)