[English] 日本語
 Yorodumi
Yorodumi- EMDB-73957: Destabilized open state sheep connexin-46/50 in DMPC nanodiscs at... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Destabilized open state sheep connexin-46/50 in DMPC nanodiscs at low pH | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | connexin / gap junction / cryo-EM / pH regulation / lipid gating / large-pore channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationgap junction-mediated intercellular transport / gap junction hemi-channel activity / connexin complex / gap junction channel activity / visual perception / cell-cell signaling / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Jarodsky JM / Myers JB / Reichow SL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2026 Journal: Nat Commun / Year: 2026Title: Reversible lipid-mediated pH-gating of connexin-46/50 by cryo-EM. Authors: Joshua M Jarodsky / Janette B Myers / Steve L Reichow /  Abstract: Gap junctions, formed by connexin proteins, establish direct electrical and metabolic coupling between cells, enabling coordinated tissue responses. These channels universally respond to ...Gap junctions, formed by connexin proteins, establish direct electrical and metabolic coupling between cells, enabling coordinated tissue responses. These channels universally respond to intracellular pH changes, closing under acidic conditions to limit the spread of cytotoxic signals during cellular stress, such as ischemia. Using cryo-electron microscopy (cryo-EM), we uncover insights into the structural mechanism of pH-gating in native lens connexin-46/50 (Cx46/50) gap junctions. Mild acidification drives lipid infiltration into the channel pore, displacing the N-terminal (NT) domain and stabilizing pore closure. Lipid involvement is shown to be both essential and fully reversible. Structural transitions involve an ensemble of gated states formed through non-cooperative NT domain movement as well as minor populations of a distinct destabilized open-state. These findings provide molecular insights into pH-gating dynamics, illustrating how structural changes may regulate gap junction function under cellular stress and linking Cx46/50 dysregulation to age-related cataract formation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_73957.map.gz emd_73957.map.gz | 8.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-73957-v30.xml emd-73957-v30.xml emd-73957.xml emd-73957.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_73957.png emd_73957.png | 132.4 KB | ||
| Filedesc metadata |  emd-73957.cif.gz emd-73957.cif.gz | 6.2 KB | ||
| Others |  emd_73957_additional_1.map.gz emd_73957_additional_1.map.gz emd_73957_half_map_1.map.gz emd_73957_half_map_1.map.gz emd_73957_half_map_2.map.gz emd_73957_half_map_2.map.gz | 106.7 MB 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-73957 http://ftp.pdbj.org/pub/emdb/structures/EMD-73957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-73957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-73957 | HTTPS FTP |
-Related structure data
| Related structure data |  9z9sMC  9z9wMC  9z7pC  9z7wC  9z81C  9z82C  9z8fC  9z8lC  9z8mC  9z9bC  9z9gC  9z9hC  9z9xC  9z9yC  9za3C  9za4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_73957.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_73957.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8016 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_73957_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_73957_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_73957_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Destabilized open state sheep connexin-46 in DMPC nanodiscs at low pH
| Entire | Name: Destabilized open state sheep connexin-46 in DMPC nanodiscs at low pH |
|---|---|
| Components |
|
-Supramolecule #1: Destabilized open state sheep connexin-46 in DMPC nanodiscs at low pH
| Supramolecule | Name: Destabilized open state sheep connexin-46 in DMPC nanodiscs at low pH type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Connexin-46 and connexin-50 coassemble in the gap junction complex and arrangement cannot be disentangled. Thus both isoforms are individually modeled into the same gap junction density. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Gap junction alpha-3 protein
| Macromolecule | Name: Gap junction alpha-3 protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.033027 KDa |
| Sequence | String: MGDWSFLGRL LENAQEHSTV IGKVWLTVLF IFRILVLGAA AEEVWGDEQS DFTCNTQQPG CENVCYDRAF PISHVRFWVL QIIFVSTPT LIYLGHVLHL VRMEEKRKER EEEPPKAAGP AEEHQDPAPV RDDRGKVRIA GALLRTYVFN IIFKTLFEVG F IAGQYFLY ...String: MGDWSFLGRL LENAQEHSTV IGKVWLTVLF IFRILVLGAA AEEVWGDEQS DFTCNTQQPG CENVCYDRAF PISHVRFWVL QIIFVSTPT LIYLGHVLHL VRMEEKRKER EEEPPKAAGP AEEHQDPAPV RDDRGKVRIA GALLRTYVFN IIFKTLFEVG F IAGQYFLY GFQLKPLYRC DRWPCPNTVD CFISRPTEKT IFILFMLAVA CVSLLLNVLE IYHLGWKKLK QGMTSPFRPD TP GSRAGSA KPMGGSPLLL PPNSAPPAVT IGFPPYYAPS ASSLGQASAP GYPEPPLPAA LPGTPGTPGT PGTLGGGGGN QGL RAPAQN CANREAEPQT SARKASPPAS TPPAAPAGGP QQFLPGGAAG SSGDSDGEGA VTAVELHAPP EPPADPGRSS KASK SSGGR ARAADLAI UniProtKB: Gap junction alpha-3 protein |
-Macromolecule #2: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 2 / Number of copies: 288 / Formula: MC3 |
|---|---|
| Molecular weight | Theoretical: 677.933 Da |
| Chemical component information | 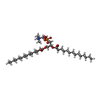 ChemComp-MC3: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 576 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 5.8 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 7072 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 29000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-9z9s:  PDB-9z9w: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































