[English] 日本語
 Yorodumi
Yorodumi- EMDB-73568: Structure of the 3-methylcrotonyl-coenzyme A carboxylase from Myc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the 3-methylcrotonyl-coenzyme A carboxylase from Mycobacterium smegmatis | |||||||||
 Map data Map data | Unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Carboxylase / transferase / leucine catabolism / metabolism / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmethylcrotonoyl-CoA carboxylase complex / methylcrotonoyl-CoA carboxylase activity / L-leucine catabolic process / biotin carboxylase / biotin carboxylase activity / transferase activity / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.0 Å | |||||||||
 Authors Authors | Liang Y / Rubinstein JL | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis for substrate specificity and MSMEG_0435-0436 binding by the mycobacterial long-chain acyl-CoA carboxylase complex Authors: Liang Y / Rubinstein JL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_73568.map.gz emd_73568.map.gz | 213.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-73568-v30.xml emd-73568-v30.xml emd-73568.xml emd-73568.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_73568_fsc.xml emd_73568_fsc.xml | 15.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_73568.png emd_73568.png | 80.4 KB | ||
| Filedesc metadata |  emd-73568.cif.gz emd-73568.cif.gz | 6.4 KB | ||
| Others |  emd_73568_additional_1.map.gz emd_73568_additional_1.map.gz emd_73568_half_map_1.map.gz emd_73568_half_map_1.map.gz emd_73568_half_map_2.map.gz emd_73568_half_map_2.map.gz | 398.8 MB 390.9 MB 390.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-73568 http://ftp.pdbj.org/pub/emdb/structures/EMD-73568 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-73568 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-73568 | HTTPS FTP |
-Related structure data
| Related structure data |  9yx0MC  9yx1C  9yx2C  9yx4C  9yx5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_73568.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_73568.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened map
| File | emd_73568_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_73568_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_73568_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 3-methylcrotonyl-coenzyme A carboxylase from Mycobacterium smegmatis
| Entire | Name: 3-methylcrotonyl-coenzyme A carboxylase from Mycobacterium smegmatis |
|---|---|
| Components |
|
-Supramolecule #1: 3-methylcrotonyl-coenzyme A carboxylase from Mycobacterium smegmatis
| Supramolecule | Name: 3-methylcrotonyl-coenzyme A carboxylase from Mycobacterium smegmatis type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 750 KDa |
-Macromolecule #1: Carboxyl transferase domain protein
| Macromolecule | Name: Carboxyl transferase domain protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 54.816199 KDa |
| Sequence | String: MSSHRDEHLA LVAELRSKLA AAALGGPERA RERHVGRGKL LPRDRVDGLL DPGSPFLELA PLAAGGMYDD ECPGAGMIAG IGRVSGREC VIVANDATVK GGTYYPITVK KHLRAQEIAL QNKLPCIYLV DSGGAFLPRQ DEVFPDRDHF GRIFYNQATM S AQGIAQIA ...String: MSSHRDEHLA LVAELRSKLA AAALGGPERA RERHVGRGKL LPRDRVDGLL DPGSPFLELA PLAAGGMYDD ECPGAGMIAG IGRVSGREC VIVANDATVK GGTYYPITVK KHLRAQEIAL QNKLPCIYLV DSGGAFLPRQ DEVFPDRDHF GRIFYNQATM S AQGIAQIA AVLGSCTAGG AYVPAMSDEA VIVRNQGTIF LGGPPLVKAA TGEVVTAEEL GGGDLHSKTS GVTDHLAHDD RD ALRIVRN IVATLGPAEP PPWQVLPAVD PIADQTELYD VVPVDARVPY DVHEVITRIV DGGEFGEFKA EYGTTLVTGF ARI HGHPVG IIANNGVLFG ESAVKGAHFI ELCDKRKTPL LFLQNISGFM VGRDYEAGGI AKHGAKMVTA VACARVPKLT VVIG GSYGA GNYSMCGRAY SPRFLWMWPN ARISVMGGEQ AASVLATVRG EMTDAEAEEF KAPIREQYEH QGNPYYSTAR LWDDG VIDP ADTRTVVGLA LSVVGQAPLE PVSYGVFRM UniProtKB: Carboxyl transferase domain protein |
-Macromolecule #2: biotin carboxylase
| Macromolecule | Name: biotin carboxylase / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO / EC number: biotin carboxylase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 70.144711 KDa |
| Sequence | String: MMMSSQTFDT VLVANRGEIA VRVIRTLRAM GIRSVAVFSE ADAGARHVTE ADVAVCIGPA AARHSYLDID AVVGAARRTD AQAVHPGYG FLSENAEFAS ALATAGIVFI GPPASAIATM GDKIAAKAAV SAFGVPVVPG ISRPGLADDD LIAGAAEVGY P VLVKPSAG ...String: MMMSSQTFDT VLVANRGEIA VRVIRTLRAM GIRSVAVFSE ADAGARHVTE ADVAVCIGPA AARHSYLDID AVVGAARRTD AQAVHPGYG FLSENAEFAS ALATAGIVFI GPPASAIATM GDKIAAKAAV SAFGVPVVPG ISRPGLADDD LIAGAAEVGY P VLVKPSAG GGGKGMRVVE AAADLPAALV SARREAGAAF GDDTLFLERF VQRPRHIEVQ VLADGHGNVI HLGERECSLQ RR HQKVIEE APSPLLDEAT RARIGAAACA TARSVDYTGA GTVEFIVSAD RPDEFFFMEM NTRLQVEHPV TELVTGIDLV EQQ IRIAAG EPLAIGQDDI TLTGHAVEAR VYAEDPAAGF LPTGGDVLGL REPTGRGVRV DSGLAAGTVV GSDYDPMLSK IIAH GSDRA SALQILDRAL ADTAVLGVTT NIEFLRFLLA DDDVAAGRLD TGLLDRRAPD FAPATVGDEQ LIAAAAYLWA RQWSA AGGD LWRVPSGWRV GEWAPATFRL HAGDRTDHVY ITGNPERASA AVEHGDTHTV SADFTPGSDT FAVTLDGLRT DYRVAV TDS QIWLSGGGRT WSVQKVREEP VRPDDAHSGD AELVSPMPGS VVAVGVPDGS DVTAGTVVVT VEAMKMEHAL TAPVDGV AK ILVAVGDQVK VGQPLARITA HTQENES UniProtKB: biotin carboxylase |
-Macromolecule #3: BIOTIN
| Macromolecule | Name: BIOTIN / type: ligand / ID: 3 / Number of copies: 6 / Formula: BTN |
|---|---|
| Molecular weight | Theoretical: 244.311 Da |
| Chemical component information | 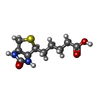 ChemComp-BTN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 35 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Number real images: 8000 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































