[English] 日本語
 Yorodumi
Yorodumi- EMDB-73575: Structure of the long chain acyl-CoA carboxylase complex from Myc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the long chain acyl-CoA carboxylase complex from Mycobacterium smegmatis | |||||||||
 Map data Map data | Structure of the long chain acyl-CoA carboxylase complex from Mycobacterium smegmatis - Composite Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Carboxylase / transferase / lipid synthesis / mycolic acid synthesis / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpropionyl-CoA carboxylase / biotin carboxylase / propionyl-CoA carboxylase activity / acetyl-CoA carboxylase complex / biotin carboxylase activity / acetyl-CoA carboxylase activity / carbon fixation / fatty acid biosynthetic process / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Liang Y / Rubinstein JL | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis for substrate specificity and MSMEG_0435-0436 binding by the mycobacterial long-chain acyl-CoA carboxylase complex Authors: Liang Y / Rubinstein JL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_73575.map.gz emd_73575.map.gz | 196.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-73575-v30.xml emd-73575-v30.xml emd-73575.xml emd-73575.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_73575.png emd_73575.png | 75.1 KB | ||
| Filedesc metadata |  emd-73575.cif.gz emd-73575.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-73575 http://ftp.pdbj.org/pub/emdb/structures/EMD-73575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-73575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-73575 | HTTPS FTP |
-Validation report
| Summary document |  emd_73575_validation.pdf.gz emd_73575_validation.pdf.gz | 571.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_73575_full_validation.pdf.gz emd_73575_full_validation.pdf.gz | 570.9 KB | Display | |
| Data in XML |  emd_73575_validation.xml.gz emd_73575_validation.xml.gz | 7.7 KB | Display | |
| Data in CIF |  emd_73575_validation.cif.gz emd_73575_validation.cif.gz | 9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-73575 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-73575 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-73575 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-73575 | HTTPS FTP |
-Related structure data
| Related structure data |  9yx1MC  9yx0C  9yx2C  9yx4C  9yx5C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_73575.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_73575.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the long chain acyl-CoA carboxylase complex from Mycobacterium smegmatis - Composite Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Long chain acyl-CoA carboxylase complex from Mycobacterium smegmatis
| Entire | Name: Long chain acyl-CoA carboxylase complex from Mycobacterium smegmatis |
|---|---|
| Components |
|
-Supramolecule #1: Long chain acyl-CoA carboxylase complex from Mycobacterium smegmatis
| Supramolecule | Name: Long chain acyl-CoA carboxylase complex from Mycobacterium smegmatis type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 870 KDa |
-Macromolecule #1: Propionyl-CoA carboxylase beta chain
| Macromolecule | Name: Propionyl-CoA carboxylase beta chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: propionyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 56.23407 KDa |
| Sequence | String: MTNKTTAELL AELREKLELA KEPGGEKAVA KREKKGIPSA RARINALLDP GSFIEIGALA KTPGDPNALY GDGVVTGRGT IDGRPVGVF SHDQTVFQGS VGEMFGRKVA RLMEWVAMVG CPIIGINDSA GARIQDAVTS LAWYAELGRR HEMLRGLVPE I SLIFGKCA ...String: MTNKTTAELL AELREKLELA KEPGGEKAVA KREKKGIPSA RARINALLDP GSFIEIGALA KTPGDPNALY GDGVVTGRGT IDGRPVGVF SHDQTVFQGS VGEMFGRKVA RLMEWVAMVG CPIIGINDSA GARIQDAVTS LAWYAELGRR HEMLRGLVPE I SLIFGKCA GGAVYSPIQT DLLVAVRDQG YMFITGPDVI KDVTGEDVTF DELGGADEQA KRGNIHKVVN SEAEAYQYVR DY LSFLPSN HFDNPPIVNP GMEPEITPHD LELDSIVPDA DNMAYDMHEI LLRIFDDGDV FEIAEQRGPA MITAFARVDG HPV GVIANQ PMVLSGAIDN EASDKAASFI RFCDSYNLPL VFVVDTPGAM PGVAEEKGGI IKRGGRFFNA IVEADVPKVT VIIR KAYGG GYAVMGSKQL SADLNFAWPT ARIAVIGAEG AAQLLVKRFP DPNAPEVQKI RDDFIEGYNL NMATPWIAAE RGYID AVIQ PHETRLLLRK SLRLLRDKQN GPKVQRKHGL LPL UniProtKB: Propionyl-CoA carboxylase beta chain |
-Macromolecule #2: Propionyl-CoA carboxylase beta chain
| Macromolecule | Name: Propionyl-CoA carboxylase beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: propionyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 58.488074 KDa |
| Sequence | String: MTSVTEPSAE HQVDIHTTAG KLADLKRRTE ETLHPVGEAA VDKVHAKGKL TARERILALL DEGSFVELDA LAKHRSTNFG LEKNRPLGD GVITGYGTID GRDVCIFSQD ATVFGGSLGE VYGEKIVKVQ ELAIKTGRPL IGINDGAGAR IQEGVVSLGL Y SRIFHNNI ...String: MTSVTEPSAE HQVDIHTTAG KLADLKRRTE ETLHPVGEAA VDKVHAKGKL TARERILALL DEGSFVELDA LAKHRSTNFG LEKNRPLGD GVITGYGTID GRDVCIFSQD ATVFGGSLGE VYGEKIVKVQ ELAIKTGRPL IGINDGAGAR IQEGVVSLGL Y SRIFHNNI KASGVIPQIS LIMGAAAGGH VYSPALTDFV VMVDQTSQMF ITGPDVIKTV TGEDVTMEEL GGAHTHMAKS GT AHYVASG EQDAFDYVRD LLSYLPPNNY ADPPLYPVAI PEGSIEETLT DEDLELDTLI PDSPNQPYDM HEVITRILDD DEF LEVQAG YAGNIVVGFG RVEGRPVGIV ANQPTQFAGC LDINASEKAA RFIRTCDCFN IPIVLLVDVP GFLPGTDQEY NGII RRGAK LLYAYGEATV AKVTVITRKS YGGAYCVMGS KDMGADVVVA WPTAQIAVMG ASGAVGFVYR QQLKEAAKNG EDVDA LRLE LQQTYEDTLV NPYIAAERGY VDAVIPPSHT RGYVANALRL LERKIVQMPP KKHGNIPL UniProtKB: Propionyl-CoA carboxylase beta chain |
-Macromolecule #3: Biotin-dependent acyl-coenzyme A carboxylase alpha3 subunit
| Macromolecule | Name: Biotin-dependent acyl-coenzyme A carboxylase alpha3 subunit type: protein_or_peptide / ID: 3 / Details: Biotin carboxyl carrier protein fragment only / Number of copies: 12 / Enantiomer: LEVO / EC number: biotin carboxylase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 63.215176 KDa |
| Sequence | String: MANHASSKIS KVLVANRGEI AVRVIRAAKD AGLASVAVYA EPDADAPHVR LADEAFALGG QTSAESYLVF EKILDAAEKS GANAIHPGY GFLSENADFA QAVIDAGLIW IGPSPQSIRD LGDKVTARHI AARAKAPLVP GTPDPVKDAD EVVAFAKEHG V PVAIKAAF ...String: MANHASSKIS KVLVANRGEI AVRVIRAAKD AGLASVAVYA EPDADAPHVR LADEAFALGG QTSAESYLVF EKILDAAEKS GANAIHPGY GFLSENADFA QAVIDAGLIW IGPSPQSIRD LGDKVTARHI AARAKAPLVP GTPDPVKDAD EVVAFAKEHG V PVAIKAAF GGGGRGMKVA RTLEEIPELF ESATREAIAA FGRGECFVER YLDKPRHVEA QVIADQHGNV VVAGTRDCSL QR RFQKLVE EAPAPFLTDA QRKEIHESAK RICKEAGYYG AGTVEYLVGQ DGLISFLEVN TRLQVEHPVT EETSGIDLVR QQF KIANGE PLDITEDPTP RGHSFEFRIN GEDAGRGFLP APGPVTKFVA PTGPGVRMDS GVETGSVIGG QFDSMLAKLI VTGA TREEA LERSRRALAE FTVEGLATVI PFHRAVVSDP AFIGDGEKFD VHTRWIETEW NNTVEPFTGG DPIEEEDTVP RQTVV VEVG GRRLEVSLPG DLAIGGGGGA AAPGVVRKKP KPRKRGGGGA KAASGDAVTA PMQGTVVKVA VEEGQEVSAG DLVVVL EAM KMENPVTAHK DGTITGLAVE AGAAITQGTV IAEIK UniProtKB: Biotin-dependent acyl-coenzyme A carboxylase alpha3 subunit |
-Macromolecule #4: Acetyl-/propionyl-coenzyme A carboxylase AccE5
| Macromolecule | Name: Acetyl-/propionyl-coenzyme A carboxylase AccE5 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 10.357693 KDa |
| Sequence | String: MSGANDSTTV SGEVQADVTD EAKADHEAHI KVLRGEPTPE EMAALMAVLA SAGGGPAEPV KKERNMWGHP VDKLRYSVFS WQRVTLLER THMRR UniProtKB: Acetyl-/propionyl-coenzyme A carboxylase AccE5 |
-Macromolecule #5: BIOTIN
| Macromolecule | Name: BIOTIN / type: ligand / ID: 5 / Number of copies: 6 / Formula: BTN |
|---|---|
| Molecular weight | Theoretical: 244.311 Da |
| Chemical component information | 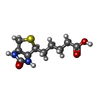 ChemComp-BTN: |
-Macromolecule #6: BICARBONATE ION
| Macromolecule | Name: BICARBONATE ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: BCT |
|---|---|
| Molecular weight | Theoretical: 61.017 Da |
| Chemical component information |  ChemComp-BCT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 35 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Number real images: 8000 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















