+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MprF from Pseudomonas aeruginosa in GDN micelle, C2 symmetry | |||||||||
 Map data Map data | Sharpened Full Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Multiple peptide resistance factor / Membrane Enzyme / Synthase / Flippase / aminoacylated lipid / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphatidylglycerol alanyltransferase activity / lysyltransferase / phosphatidylglycerol lysyltransferase activity / phospholipid homeostasis / lipid metabolic process / response to antibiotic / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Jha S / Vinothkumar KR | |||||||||
| Funding support |  India, 2 items India, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structures of Multiple Peptide Resistance Factor from Pseudomonas aeruginosa Authors: Jha S / Vinothkumar KR | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61238.map.gz emd_61238.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61238-v30.xml emd-61238-v30.xml emd-61238.xml emd-61238.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61238_fsc.xml emd_61238_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_61238.png emd_61238.png | 110.2 KB | ||
| Filedesc metadata |  emd-61238.cif.gz emd-61238.cif.gz | 7.8 KB | ||
| Others |  emd_61238_half_map_1.map.gz emd_61238_half_map_1.map.gz emd_61238_half_map_2.map.gz emd_61238_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61238 http://ftp.pdbj.org/pub/emdb/structures/EMD-61238 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61238 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61238 | HTTPS FTP |
-Related structure data
| Related structure data |  9j8qMC  9j8rC  9j8sC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61238.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61238.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened Full Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Unsharpened Half Map
| File | emd_61238_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened Half Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unsharpened Half Map
| File | emd_61238_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened Half Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MprF dimer
| Entire | Name: MprF dimer |
|---|---|
| Components |
|
-Supramolecule #1: MprF dimer
| Supramolecule | Name: MprF dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) / Location in cell: Cell membrane Pseudomonas aeruginosa PAO1 (bacteria) / Location in cell: Cell membrane |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Phosphatidylglycerol lysyltransferase
| Macromolecule | Name: Phosphatidylglycerol lysyltransferase / type: protein_or_peptide / ID: 1 Details: 1-9: MWSHPQFEK - (Strep Tag II), 10-13: GGSG - (Linker), 14-894: PaMprF, 895-899 (RNSSS) - additional residues, from fusion protein 900-903 - VDAL - (N terminus of CPD after cleavage), 1-32, ...Details: 1-9: MWSHPQFEK - (Strep Tag II), 10-13: GGSG - (Linker), 14-894: PaMprF, 895-899 (RNSSS) - additional residues, from fusion protein 900-903 - VDAL - (N terminus of CPD after cleavage), 1-32, 713-726, 805-832, 873-881 - Not modelled (numbering based on Uniprot and doesn't include the fusion construct) Number of copies: 2 / Enantiomer: LEVO / EC number: lysyltransferase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 |
| Molecular weight | Theoretical: 98.330453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MWSHPQFEKG GSGMRTDAPV PEHPAPPSSP ASPQRIRLID RITAYRQPIG LVFTLLLFGL ALVACYHLLR EIDPGALHDA IADVPRPAL LGALSATALG FVILLGYEWS ASRFAGVTLP MRSLATGGFS AFAIGNAVGL SLLSGGSVRY RLYSRHGIGA A EIARMTLF ...String: MWSHPQFEKG GSGMRTDAPV PEHPAPPSSP ASPQRIRLID RITAYRQPIG LVFTLLLFGL ALVACYHLLR EIDPGALHDA IADVPRPAL LGALSATALG FVILLGYEWS ASRFAGVTLP MRSLATGGFS AFAIGNAVGL SLLSGGSVRY RLYSRHGIGA A EIARMTLF ASLSLGCALP VLAALAALCD LDDAASALHL PRALVAVIAI AVLSLAVGLV AFLARHRLPG ERPSPDSLLV RL GRRSLRL PGLRLSLLQL LITALDVAAA ATVLYLLLPE TPPFAAFLLV YLLALAAGVL SHVPGGVGVF EAVLLAAFAG QLG AAPLAA ALLLYRLIYV VLPLLLACLL LLFLEARRLW VTRQAIRVAS GFAAPILAIL VFLSGVVLLF SGATPAIDTR LEHL GFLIP HRLIDASHLV ASLIGVLCLL LAQGLRRRLS AAWALTLVLL LVGALLSLLK GFDWEEASLL SLTAALLAMF RRSFY RPSR LMEVPFSPLY VGASICVVGA SVWLLLFANQ DVHYSNQLWW QFALDADAPR ALRAALGSCL LLLALALGWL LRAAPP AIR EPNAEELQRA ARIIRHSDQP DGGLALTGDK ALLFHESDDA FLMYARRGRS MIALYDPIGP AMQRAELIWQ FRDLCDL HH ARPVFYQVRA ENLPFYMDIG LTALKLGEEA RVDLLRFDLE NKGKEMKDLR YTWNRGQRDG LALEFHEPGQ APLDELKA I SDAWLGGKQV REKGFSLGRF TPAYLNFFRI AIVRHQGKPV AFANLLETDS RELASLDLMR VHPDAPKLTM EFLMLGLIL HYKAQGHARF SLGMVPLAGL QPRRGAPLTQ RLGALVFRRG EQFYNFQGLR RFKDKFQPDW EPRYLAVPAG LDPLVALADT AALIAGGLT GLVKRRNSSS VDAL UniProtKB: Phosphatidylglycerol lysyltransferase |
-Macromolecule #2: (2~{R},3~{S},4~{S},5~{S},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{S}...
| Macromolecule | Name: (2~{R},3~{S},4~{S},5~{S},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{S},4~{R},5~{R},6~{R})-2-(hydroxymethyl)-6-[2-[[(2~{R},3~{S},4~{R},5~{R},6~{S})-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{S},6~{R}) ...Name: (2~{R},3~{S},4~{S},5~{S},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{S},4~{R},5~{R},6~{R})-2-(hydroxymethyl)-6-[2-[[(2~{R},3~{S},4~{R},5~{R},6~{S})-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]oxymethyl]-4-[(1~{R},2~{R},4~{S},5'~{R},6~{R},7~{R},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxy-butoxy]-4,5-bis(oxidanyl)oxan-3-yl]oxy-oxane-3,4,5-triol type: ligand / ID: 2 / Number of copies: 2 / Formula: J4U |
|---|---|
| Molecular weight | Theoretical: 1.165315 KDa |
| Chemical component information | 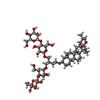 ChemComp-J4U: |
-Macromolecule #3: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 3 / Number of copies: 22 / Formula: PGT |
|---|---|
| Molecular weight | Theoretical: 751.023 Da |
| Chemical component information |  ChemComp-PGT: |
-Macromolecule #4: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 4 / Number of copies: 12 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR Details: The grid was glow discharged at 25 mA with PELCO easyglow | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force, 0. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 2 / Number real images: 3877 / Average exposure time: 60.0 sec. / Average electron dose: 28.75 e/Å2 Details: Images were collected in movie-mode at 40 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 13084 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 146.5 | |||||||||
| Output model |  PDB-9j8q: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






































