[English] 日本語
 Yorodumi
Yorodumi- EMDB-60562: Cryo-EM structure of uropathogenic Escherichia coli CysK:CdiA:tRN... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of uropathogenic Escherichia coli CysK:CdiA:tRNA complex B | |||||||||
 Map data Map data | Postprocessed by deepEMhancer. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNase / Complex / Contact-dependent growth inhibition / Toxin | |||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA-specific ribonuclease activity / cysteine synthase / L-cysteine desulfhydrase activity / cysteine synthase activity / other organism cell membrane / cysteine biosynthetic process from serine / RNA endonuclease activity / toxin activity / host cell cytoplasm / tRNA binding ...tRNA-specific ribonuclease activity / cysteine synthase / L-cysteine desulfhydrase activity / cysteine synthase activity / other organism cell membrane / cysteine biosynthetic process from serine / RNA endonuclease activity / toxin activity / host cell cytoplasm / tRNA binding / Hydrolases; Acting on ester bonds / extracellular region / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
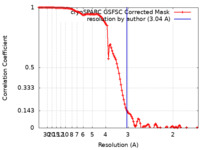
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Feng Z / Yashiro Y / Tomita K | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Mechanism of activation of contact-dependent growth inhibition tRNase toxin by the amino acid biogenesis factor CysK in the bacterial competition system. Authors: Feng Z / Yashiro Y / Tomita K | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60562.map.gz emd_60562.map.gz | 126.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60562-v30.xml emd-60562-v30.xml emd-60562.xml emd-60562.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60562_fsc.xml emd_60562_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_60562.png emd_60562.png | 83 KB | ||
| Masks |  emd_60562_msk_1.map emd_60562_msk_1.map | 144.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-60562.cif.gz emd-60562.cif.gz | 6.9 KB | ||
| Others |  emd_60562_half_map_1.map.gz emd_60562_half_map_1.map.gz emd_60562_half_map_2.map.gz emd_60562_half_map_2.map.gz | 134.5 MB 134.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60562 http://ftp.pdbj.org/pub/emdb/structures/EMD-60562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60562 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60562 | HTTPS FTP |
-Validation report
| Summary document |  emd_60562_validation.pdf.gz emd_60562_validation.pdf.gz | 815 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60562_full_validation.pdf.gz emd_60562_full_validation.pdf.gz | 814.6 KB | Display | |
| Data in XML |  emd_60562_validation.xml.gz emd_60562_validation.xml.gz | 19.7 KB | Display | |
| Data in CIF |  emd_60562_validation.cif.gz emd_60562_validation.cif.gz | 25.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60562 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60562 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60562 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60562 | HTTPS FTP |
-Related structure data
| Related structure data |  8zydMC  8zycC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60562.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60562.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed by deepEMhancer. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
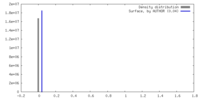
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_60562_msk_1.map emd_60562_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
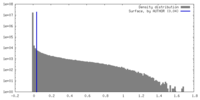
| Density Histograms |
-Half map: #2
| File | emd_60562_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
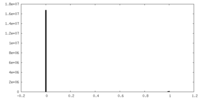
| Density Histograms |
-Half map: #1
| File | emd_60562_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
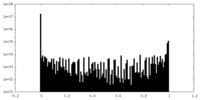
| Density Histograms |
- Sample components
Sample components
-Entire : CysK in complex with CdiA-CT and tRNA.
| Entire | Name: CysK in complex with CdiA-CT and tRNA. |
|---|---|
| Components |
|
-Supramolecule #1: CysK in complex with CdiA-CT and tRNA.
| Supramolecule | Name: CysK in complex with CdiA-CT and tRNA. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 110 KDa |
-Macromolecule #1: Cysteine synthase A
| Macromolecule | Name: Cysteine synthase A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: cysteine synthase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.756688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKIFEDNSL TIGHTPLVRL NRIGNGRILA KVESRNPSFS V(LLP)CRIGANMI WDAEKRGVLK PGVELVEPTS GNTGIA LAY VAAARGYKLT LTMPETMSIE RRKLLKALGA NLVLTEGAKG MKGAIQKAEE IVASNPEKYL LLQQFSNPAN PEIHEKT TG PEIWEDTDGQ ...String: MSKIFEDNSL TIGHTPLVRL NRIGNGRILA KVESRNPSFS V(LLP)CRIGANMI WDAEKRGVLK PGVELVEPTS GNTGIA LAY VAAARGYKLT LTMPETMSIE RRKLLKALGA NLVLTEGAKG MKGAIQKAEE IVASNPEKYL LLQQFSNPAN PEIHEKT TG PEIWEDTDGQ VDVFIAGVGT GGTLTGVSRY IKGTKGKTDL ISVAVEPTDS PVIAQALAGE EIKPGPHKIQ GIGAGFIP A NLDLKLVDKV IGITNEEAIS TARRLMEEEG ILAGISSGAA VAAALKLQED ESFTNKNIVV ILPSSGERYL STALFADLF TEKELQQ UniProtKB: Cysteine synthase A |
-Macromolecule #2: tRNA nuclease CdiA
| Macromolecule | Name: tRNA nuclease CdiA / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.18125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHVEN NALSLVARGC AVAAPCRTKV AEQLLEIGAK AGMAGLAGAA VKDMADRMTS DELEHLITLQ MMGNDEITTK YLSSLHDKY GSGAASNPNI GKDLTDAEKV ELGGSGSGTG TPPPSENDPK QQNEKTVDKL NQKQESAIKK IDNTIKNALK D HDIIGTLK ...String: MHHHHHHVEN NALSLVARGC AVAAPCRTKV AEQLLEIGAK AGMAGLAGAA VKDMADRMTS DELEHLITLQ MMGNDEITTK YLSSLHDKY GSGAASNPNI GKDLTDAEKV ELGGSGSGTG TPPPSENDPK QQNEKTVDKL NQKQESAIKK IDNTIKNALK D HDIIGTLK DMDGKPVPKE NGGYWDAMQE MQNTLRGLRN HADTLKNVNN PEAQAAYGRA TDAINKIESA LKGYGI UniProtKB: tRNA nuclease CdiA |
-Macromolecule #3: tRNAIleGAU
| Macromolecule | Name: tRNAIleGAU / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.000869 KDa |
| Sequence | String: AGGCUUGUAG CUCAGGUGGU UAGAGCGCAC CCCUGAU(T6A)AG GGUGAGGUCG GUGGUUCAAG UCCACUCAGG CCUACC A GENBANK: GENBANK: CP053605.1 |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
Details: 25mM Tris-HCl,50mM NaCl,2mM MgCl2, 10mM 2-mercaptoethanol | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 4092 pixel / Digitization - Dimensions - Height: 5760 pixel / Number grids imaged: 1 / Number real images: 7044 / Average exposure time: 1.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)