+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6000 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full virus map of Brome Mosaic Virus | |||||||||
 Map data Map data | full BMV map with combined data | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Bromovirus coat protein / Bromovirus coat protein / T=3 icosahedral viral capsid / host cell endoplasmic reticulum / viral nucleocapsid / ribonucleoprotein complex / structural molecule activity / RNA binding / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   Brome mosaic virus Brome mosaic virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Wang Z / Hryc FC / Bammes B / Afonine PV / Jakana J / Chen DH / Liu XA / Baker ML / Kao C / Ludtke SJ ...Wang Z / Hryc FC / Bammes B / Afonine PV / Jakana J / Chen DH / Liu XA / Baker ML / Kao C / Ludtke SJ / Schmid MF / Adams PD / Chiu W | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2014 Journal: Nat Commun / Year: 2014Title: An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Authors: Zhao Wang / Corey F Hryc / Benjamin Bammes / Pavel V Afonine / Joanita Jakana / Dong-Hua Chen / Xiangan Liu / Matthew L Baker / Cheng Kao / Steven J Ludtke / Michael F Schmid / Paul D Adams / Wah Chiu /  Abstract: Advances in electron cryo-microscopy have enabled structure determination of macromolecules at near-atomic resolution. However, structure determination, even using de novo methods, remains ...Advances in electron cryo-microscopy have enabled structure determination of macromolecules at near-atomic resolution. However, structure determination, even using de novo methods, remains susceptible to model bias and overfitting. Here we describe a complete workflow for data acquisition, image processing, all-atom modelling and validation of brome mosaic virus, an RNA virus. Data were collected with a direct electron detector in integrating mode and an exposure beyond the traditional radiation damage limit. The final density map has a resolution of 3.8 Å as assessed by two independent data sets and maps. We used the map to derive an all-atom model with a newly implemented real-space optimization protocol. The validity of the model was verified by its match with the density map and a previous model from X-ray crystallography, as well as the internal consistency of models from independent maps. This study demonstrates a practical approach to obtain a rigorously validated atomic resolution electron cryo-microscopy structure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6000.map.gz emd_6000.map.gz | 203 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6000-v30.xml emd-6000-v30.xml emd-6000.xml emd-6000.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-6000_image.png EMD-6000_image.png | 967.4 KB | ||
| Others |  emd_6000_additional_1.map.gz emd_6000_additional_1.map.gz emd_6000_additional_2.map.gz emd_6000_additional_2.map.gz emd_6000_additional_3.map.gz emd_6000_additional_3.map.gz emd_6000_additional_4.map.gz emd_6000_additional_4.map.gz emd_6000_additional_5.map.gz emd_6000_additional_5.map.gz | 392 KB 404.3 KB 404.5 KB 229.9 MB 229.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6000 http://ftp.pdbj.org/pub/emdb/structures/EMD-6000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6000 | HTTPS FTP |
-Related structure data
| Related structure data |  3j7lMC  3j7mMC  3j7nMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10010 (Title: Full virus map of Brome Mosaic Virus (micrographs and particle coordinates) EMPIAR-10010 (Title: Full virus map of Brome Mosaic Virus (micrographs and particle coordinates)Data size: 1.7 TB Data #1: Brome Mosaic Virus micrographs - non gain corrected [micrographs - multiframe] Data #2: Brome Mosaic Virus micrographs - gain corrected [micrographs - multiframe])  EMPIAR-10011 (Title: Full virus map of Brome Mosaic Virus (picked particles) EMPIAR-10011 (Title: Full virus map of Brome Mosaic Virus (picked particles)Data size: 23.1 Data #1: Brome Mosaic Virus boxed particles [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6000.map.gz / Format: CCP4 / Size: 276 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6000.map.gz / Format: CCP4 / Size: 276 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full BMV map with combined data | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
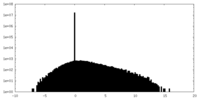
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.99 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
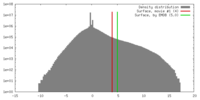
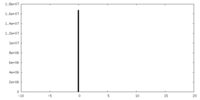
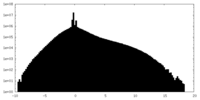
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 6000 additional 1.map
| File | emd_6000_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
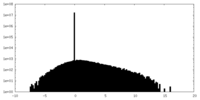
| Density Histograms |
-Supplemental map: emd 6000 additional 2.map
| File | emd_6000_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
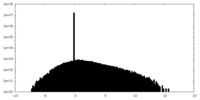
| Projections & Slices |
| ||||||||||||
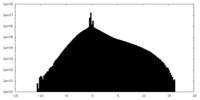
| Density Histograms |
-Supplemental map: emd 6000 additional 3.map
| File | emd_6000_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
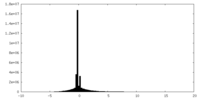
| Density Histograms |
-Supplemental map: emd 6000 additional 4.map
| File | emd_6000_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 6000 additional 5.map
| File | emd_6000_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : brome mosaic virus
| Entire | Name:   brome mosaic virus brome mosaic virus |
|---|---|
| Components |
|
-Supramolecule #1000: brome mosaic virus
| Supramolecule | Name: brome mosaic virus / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 4.6 MDa |
-Supramolecule #1: Brome mosaic virus
| Supramolecule | Name: Brome mosaic virus / type: virus / ID: 1 / NCBI-ID: 12302 / Sci species name: Brome mosaic virus / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 171.0 K / Instrument: FEI VITROBOT MARK IV |
|---|
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Date | Jan 10, 2013 |
| Image recording | Category: CCD / Film or detector model: DIRECT ELECTRON DE-12 (4k x 3k) |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: OTHER / Software - Name: MPSA, EMAN / Number images used: 30000 |
|---|
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)