[English] 日本語
 Yorodumi
Yorodumi- EMDB-5941: Cryo-EM structure of the small subunit of the mammalian mitochond... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5941 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome | |||||||||
 Map data Map data | mammalian mito-ribosome small subunit structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mammalian mito-ribosome small subunit structure | |||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial translation elongation / Mitochondrial translation termination / peptide biosynthetic process / mitochondrial ribosome assembly / mitochondrial ribosome / mitochondrial small ribosomal subunit / mitochondrial translation / Mitochondrial protein degradation / ribosomal small subunit binding / kinase activity ...Mitochondrial translation elongation / Mitochondrial translation termination / peptide biosynthetic process / mitochondrial ribosome assembly / mitochondrial ribosome / mitochondrial small ribosomal subunit / mitochondrial translation / Mitochondrial protein degradation / ribosomal small subunit binding / kinase activity / regulation of translation / ribosomal small subunit assembly / small ribosomal subunit rRNA binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / tRNA binding / cell population proliferation / mitochondrial inner membrane / rRNA binding / ribosome / structural constituent of ribosome / protein domain specific binding / translation / ribonucleoprotein complex / mRNA binding / apoptotic process / nucleolus / mitochondrion / RNA binding / nucleoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.0 Å | |||||||||
 Authors Authors | Kaushal PS / Sharma MR / Booth TM / Haque E / Tung C / Sanbonmatsu K / Spremulli L / Agrawal RK | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2014 Journal: Proc Natl Acad Sci U S A / Year: 2014Title: Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Authors: Prem S Kaushal / Manjuli R Sharma / Timothy M Booth / Emdadul M Haque / Chang-Shung Tung / Karissa Y Sanbonmatsu / Linda L Spremulli / Rajendra K Agrawal /  Abstract: The mammalian mitochondrial ribosomes (mitoribosomes) are responsible for synthesizing 13 membrane proteins that form essential components of the complexes involved in oxidative phosphorylation or ...The mammalian mitochondrial ribosomes (mitoribosomes) are responsible for synthesizing 13 membrane proteins that form essential components of the complexes involved in oxidative phosphorylation or ATP generation for the eukaryotic cell. The mammalian 55S mitoribosome contains significantly smaller rRNAs and a large mass of mitochondrial ribosomal proteins (MRPs), including large mito-specific amino acid extensions and insertions in MRPs that are homologous to bacterial ribosomal proteins and an additional 35 mito-specific MRPs. Here we present the cryo-EM structure analysis of the small (28S) subunit (SSU) of the 55S mitoribosome. We find that the mito-specific extensions in homologous MRPs generally are involved in inter-MRP contacts and in contacts with mito-specific MRPs, suggesting a stepwise evolution of the current architecture of the mitoribosome. Although most of the mito-specific MRPs and extensions of homologous MRPs are situated on the peripheral regions, they also contribute significantly to the formation of linings of the mRNA and tRNA paths, suggesting a tailor-made structural organization of the mito-SSU for the recruitment of mito-specific mRNAs, most of which do not possess a 5' leader sequence. In addition, docking of previously published coordinates of the large (39S) subunit (LSU) into the cryo-EM map of the 55S mitoribosome reveals that mito-specific MRPs of both the SSU and LSU are involved directly in the formation of six of the 15 intersubunit bridges. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5941.map.gz emd_5941.map.gz | 186.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5941-v30.xml emd-5941-v30.xml emd-5941.xml emd-5941.xml | 11 KB 11 KB | Display Display |  EMDB header EMDB header |
| Images |  400_5941.gif 400_5941.gif 80_5941.gif 80_5941.gif | 51.9 KB 3.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5941 http://ftp.pdbj.org/pub/emdb/structures/EMD-5941 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5941 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5941 | HTTPS FTP |
-Validation report
| Summary document |  emd_5941_validation.pdf.gz emd_5941_validation.pdf.gz | 315.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5941_full_validation.pdf.gz emd_5941_full_validation.pdf.gz | 315.4 KB | Display | |
| Data in XML |  emd_5941_validation.xml.gz emd_5941_validation.xml.gz | 6.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5941 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5941 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5941 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5941 | HTTPS FTP |
-Related structure data
| Related structure data |  3jcq  3jd5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5941.map.gz / Format: CCP4 / Size: 196.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5941.map.gz / Format: CCP4 / Size: 196.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mammalian mito-ribosome small subunit structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.17 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
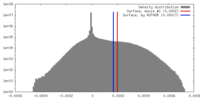
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : small subunit of mito-ribosome
| Entire | Name: small subunit of mito-ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: small subunit of mito-ribosome
| Supramolecule | Name: small subunit of mito-ribosome / type: sample / ID: 1000 / Details: monodisperse / Oligomeric state: 1 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 1.18 MDa |
-Supramolecule #1: mito-ribosomal small subunit
| Supramolecule | Name: mito-ribosomal small subunit / type: complex / ID: 1 / Name.synonym: 28S mito-ribosome / Details: Single particle Cryo-EM of mito-ribosome / Recombinant expression: No / Database: NCBI / Ribosome-details: ribosome-eukaryote: SSU 40S |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.18 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.86 mg/mL |
|---|---|
| Buffer | pH: 7.6 Details: 20 mM HEPES-KOH, pH 7.6, 20 mM MgCl2, 40 mM KCl, 20 mM DTT |
| Grid | Details: 300 mesh Quantifoil holey carbon copper grid, carbon support, glow discharged in a plasma cleaner |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 120 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FS |
|---|---|
| Temperature | Min: 80 K / Max: 105 K / Average: 100 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification |
| Date | Oct 10, 2009 |
| Image recording | Category: FILM / Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 0.5 µm / Number real images: 866553 / Average electron dose: 9 e/Å2 / Camera length: 800 / Od range: 1.4 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 59717 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 1.4 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)