+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
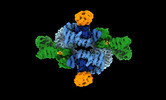
| Title | cryo-EM structure of human LST2 bound to human mTOR complex 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MTOR / MTORC1 / LST2 / ZFYVE28 / EGFR / TOS / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of epidermal growth factor-activated receptor activity / RNA polymerase III type 2 promoter sequence-specific DNA binding / RNA polymerase III type 1 promoter sequence-specific DNA binding / positive regulation of cytoplasmic translational initiation / regulation of locomotor rhythm / T-helper 1 cell lineage commitment / positive regulation of pentose-phosphate shunt / positive regulation of wound healing, spreading of epidermal cells / TORC2 signaling / TORC2 complex ...negative regulation of epidermal growth factor-activated receptor activity / RNA polymerase III type 2 promoter sequence-specific DNA binding / RNA polymerase III type 1 promoter sequence-specific DNA binding / positive regulation of cytoplasmic translational initiation / regulation of locomotor rhythm / T-helper 1 cell lineage commitment / positive regulation of pentose-phosphate shunt / positive regulation of wound healing, spreading of epidermal cells / TORC2 signaling / TORC2 complex / regulation of membrane permeability / cellular response to leucine starvation / heart valve morphogenesis / TFIIIC-class transcription factor complex binding / negative regulation of lysosome organization / TORC1 complex / voluntary musculoskeletal movement / positive regulation of transcription of nucleolar large rRNA by RNA polymerase I / positive regulation of odontoblast differentiation / calcineurin-NFAT signaling cascade / RNA polymerase III type 3 promoter sequence-specific DNA binding / positive regulation of keratinocyte migration / regulation of osteoclast differentiation / regulation of lysosome organization / MTOR signalling / cellular response to L-leucine / energy reserve metabolic process / regulation of autophagosome assembly / Energy dependent regulation of mTOR by LKB1-AMPK / cellular response to nutrient / TORC1 signaling / Amino acids regulate mTORC1 / phosphatidylinositol-3-phosphate binding / serine/threonine protein kinase complex / cellular response to methionine / ruffle organization / negative regulation of cell size / positive regulation of ubiquitin-dependent protein catabolic process / positive regulation of osteoclast differentiation / cellular response to osmotic stress / anoikis / inositol hexakisphosphate binding / negative regulation of protein localization to nucleus / cardiac muscle cell development / negative regulation of calcineurin-NFAT signaling cascade / regulation of myelination / positive regulation of transcription by RNA polymerase III / negative regulation of macroautophagy / Macroautophagy / positive regulation of myotube differentiation / regulation of cell size / Constitutive Signaling by AKT1 E17K in Cancer / negative regulation of epidermal growth factor receptor signaling pathway / positive regulation of actin filament polymerization / germ cell development / behavioral response to pain / oligodendrocyte differentiation / positive regulation of oligodendrocyte differentiation / TOR signaling / positive regulation of translational initiation / mTORC1-mediated signalling / CD28 dependent PI3K/Akt signaling / social behavior / HSF1-dependent transactivation / positive regulation of G1/S transition of mitotic cell cycle / regulation of macroautophagy / protein serine/threonine kinase inhibitor activity / protein kinase activator activity / positive regulation of TOR signaling / enzyme-substrate adaptor activity / 'de novo' pyrimidine nucleobase biosynthetic process / response to amino acid / positive regulation of epithelial to mesenchymal transition / positive regulation of lipid biosynthetic process / vascular endothelial cell response to laminar fluid shear stress / cellular response to nutrient levels / heart morphogenesis / neuronal action potential / regulation of cellular response to heat / positive regulation of lamellipodium assembly / cardiac muscle contraction / positive regulation of stress fiber assembly / T cell costimulation / phagocytic vesicle / 14-3-3 protein binding / positive regulation of endothelial cell proliferation / endomembrane system / cytoskeleton organization / negative regulation of insulin receptor signaling pathway / negative regulation of autophagy / cellular response to amino acid starvation / cellular response to starvation / positive regulation of glycolytic process / regulation of signal transduction by p53 class mediator / Regulation of PTEN gene transcription / protein serine/threonine kinase activator activity / positive regulation of translation / VEGFR2 mediated vascular permeability / post-embryonic development / regulation of actin cytoskeleton organization Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
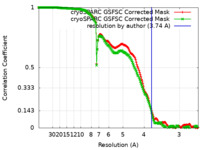
| Method | single particle reconstruction / cryo EM / Resolution: 3.74 Å | |||||||||
 Authors Authors | Craigie LM / Maier T | |||||||||
| Funding support |  Switzerland, European Union, 2 items Switzerland, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: mTORC1 phosphorylates and stabilizes LST2 to negatively regulate EGFR. Authors: Stefania Battaglioni / Louise-Marie Craigie / Sofia Filippini / Timm Maier / Michael N Hall /  Abstract: TORC1 (target of rapamycin complex 1) is a highly conserved protein kinase that plays a central role in regulating cell growth. Given the role of mammalian TORC1 (mTORC1) in metabolism and disease, ...TORC1 (target of rapamycin complex 1) is a highly conserved protein kinase that plays a central role in regulating cell growth. Given the role of mammalian TORC1 (mTORC1) in metabolism and disease, understanding mTORC1 downstream signaling and feedback loops is important. mTORC1 recognizes some of its substrates via a five amino acid binding sequence called the TOR signaling (TOS) motif. mTORC1 binding to a TOS motif facilitates phosphorylation of a distinct, distal site. Here, we show that LST2, also known as ZFYVE28, contains a TOS motif (amino acids 401 to 405) and is directly phosphorylated by mTORC1 at serine 670 (S670). mTORC1-mediated S670 phosphorylation promotes LST2 monoubiquitination on lysine 87 (K87). Monoubiquitinated LST2 is stable and displays a broad reticular distribution. When mTORC1 is inactive, unphosphorylated LST2 is degraded by the proteasome. The absence of LST2 enhances EGFR (epidermal growth factor receptor) signaling. We propose that mTORC1 negatively feeds back on its upstream receptor EGFR via LST2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50184.map.gz emd_50184.map.gz | 244.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50184-v30.xml emd-50184-v30.xml emd-50184.xml emd-50184.xml | 26.7 KB 26.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50184_fsc.xml emd_50184_fsc.xml emd_50184_fsc_2.xml emd_50184_fsc_2.xml | 16.9 KB 16.9 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_50184.png emd_50184.png | 45.5 KB | ||
| Filedesc metadata |  emd-50184.cif.gz emd-50184.cif.gz | 9.5 KB | ||
| Others |  emd_50184_additional_1.map.gz emd_50184_additional_1.map.gz emd_50184_additional_2.map.gz emd_50184_additional_2.map.gz emd_50184_additional_3.map.gz emd_50184_additional_3.map.gz | 257.4 MB 257.6 MB 257.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50184 http://ftp.pdbj.org/pub/emdb/structures/EMD-50184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50184 | HTTPS FTP |
-Related structure data
| Related structure data |  9f45MC  9f42C  9f43C  9f44C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50184.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50184.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #3
| File | emd_50184_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_50184_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_50184_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mTORC1 in complex with LST2
| Entire | Name: mTORC1 in complex with LST2 |
|---|---|
| Components |
|
-Supramolecule #1: mTORC1 in complex with LST2
| Supramolecule | Name: mTORC1 in complex with LST2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Serine/threonine-protein kinase mTOR
| Macromolecule | Name: Serine/threonine-protein kinase mTOR / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 289.257969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLGTGPAAAT TAATTSSNVS VLQQFASGLK SRNEETRAKA AKELQHYVTM ELREMSQEES TRFYDQLNHH IFELVSSSDA NERKGGILA IASLIGVEGG NATRIGRFAN YLRNLLPSND PVVMEMASKA IGRLAMAGDT FTAEYVEFEV KRALEWLGAD R NEGRRHAA ...String: MLGTGPAAAT TAATTSSNVS VLQQFASGLK SRNEETRAKA AKELQHYVTM ELREMSQEES TRFYDQLNHH IFELVSSSDA NERKGGILA IASLIGVEGG NATRIGRFAN YLRNLLPSND PVVMEMASKA IGRLAMAGDT FTAEYVEFEV KRALEWLGAD R NEGRRHAA VLVLRELAIS VPTFFFQQVQ PFFDNIFVAV WDPKQAIREG AVAALRACLI LTTQREPKEM QKPQWYRHTF EE AEKGFDE TLAKEKGMNR DDRIHGALLI LNELVRISSM EGERLREEME EITQQQLVHD KYCKDLMGFG TKPRHITPFT SFQ AVQPQQ SNALVGLLGY SSHQGLMGFG TSPSPAKSTL VESRCCRDLM EEKFDQVCQW VLKCRNSKNS LIQMTILNLL PRLA AFRPS AFTDTQYLQD TMNHVLSCVK KEKERTAAFQ ALGLLSVAVR SEFKVYLPRV LDIIRAALPP KDFAHKRQKA MQVDA TVFT CISMLARAMG PGIQQDIKEL LEPMLAVGLS PALTAVLYDL SRQIPQLKKD IQDGLLKMLS LVLMHKPLRH PGMPKG LAH QLASPGLTTL PEASDVGSIT LALRTLGSFE FEGHSLTQFV RHCADHFLNS EHKEIRMEAA RTCSRLLTPS IHLISGH AH VVSQTAVQVV ADVLSKLLVV GITDPDPDIR YCVLASLDER FDAHLAQAEN LQALFVALND QVFEIRELAI CTVGRLSS M NPAFVMPFLR KMLIQILTEL EHSGIGRIKE QSARMLGHLV SNAPRLIRPY MEPILKALIL KLKDPDPDPN PGVINNVLA TIGELAQVSG LEMRKWVDEL FIIIMDMLQD SSLLAKRQVA LWTLGQLVAS TGYVVEPYRK YPTLLEVLLN FLKTEQNQGT RREAIRVLG LLGALDPYKH KVNIGMIDQS RDASAVSLSE SKSSQDSSDY STSEMLVNMG NLPLDEFYPA VSMVALMRIF R DQSLSHHH TMVVQAITFI FKSLGLKCVQ FLPQVMPTFL NVIRVCDGAI REFLFQQLGM LVSFVKSHIR PYMDEIVTLM RE FWVMNTS IQSTIILLIE QIVVALGGEF KLYLPQLIPH MLRVFMHDNS PGRIVSIKLL AAIQLFGANL DDYLHLLLPP IVK LFDAPE APLPSRKAAL ETVDRLTESL DFTDYASRII HPIVRTLDQS PELRSTAMDT LSSLVFQLGK KYQIFIPMVN KVLV RHRIN HQRYDVLICR IVKGYTLADE EEDPLIYQHR MLRSGQGDAL ASGPVETGPM KKLHVSTINL QKAWGAARRV SKDDW LEWL RRLSLELLKD SSSPSLRSCW ALAQAYNPMA RDLFNAAFVS CWSELNEDQQ DELIRSIELA LTSQDIAEVT QTLLNL AEF MEHSDKGPLP LRDDNGIVLL GERAAKCRAY AKALHYKELE FQKGPTPAIL ESLISINNKL QQPEAAAGVL EYAMKHF GE LEIQATWYEK LHEWEDALVA YDKKMDTNKD DPELMLGRMR CLEALGEWGQ LHQQCCEKWT LVNDETQAKM ARMAAAAA W GLGQWDSMEE YTCMIPRDTH DGAFYRAVLA LHQDLFSLAQ QCIDKARDLL DAELTAMAGE SYSRAYGAMV SCHMLSELE EVIQYKLVPE RREIIRQIWW ERLQGCQRIV EDWQKILMVR SLVVSPHEDM RTWLKYASLC GKSGRLALAH KTLVLLLGVD PSRQLDHPL PTVHPQVTYA YMKNMWKSAR KIDAFQHMQH FVQTMQQQAQ HAIATEDQQH KQELHKLMAR CFLKLGEWQL N LQGINEST IPKVLQYYSA ATEHDRSWYK AWHAWAVMNF EAVLHYKHQN QARDEKKKLR HASGANITNA TTAATTAATA TT TASTEGS NSESEAESTE NSPTPSPLQK KVTEDLSKTL LMYTVPAVQG FFRSISLSRG NNLQDTLRVL TLWFDYGHWP DVN EALVEG VKAIQIDTWL QVIPQLIARI DTPRPLVGRL IHQLLTDIGR YHPQALIYPL TVASKSTTTA RHNAANKILK NMCE HSNTL VQQAMMVSEE LIRVAILWHE MWHEGLEEAS RLYFGERNVK GMFEVLEPLH AMMERGPQTL KETSFNQAYG RDLME AQEW CRKYMKSGNV KDLTQAWDLY YHVFRRISKQ LPQLTSLELQ YVSPKLLMCR DLELAVPGTY DPNQPIIRIQ SIAPSL QVI TSKQRPRKLT LMGSNGHEFV FLLKGHEDLR QDERVMQLFG LVNTLLANDP TSLRKNLSIQ RYAVIPLSTN SGLIGWV PH CDTLHALIRD YREKKKILLN IEHRIMLRMA PDYDHLTLMQ KVEVFEHAVN NTAGDDLAKL LWLKSPSSEV WFDRRTNY T RSLAVMSMVG YILGLGDRHP SNLMLDRLSG KILHIDFGDC FEVAMTREKF PEKIPFRLTR MLTNAMEVTG LDGNYRITC HTVMEVLREH KDSVMAVLEA FVYDPLLNWR LMDTNTKGNK RSRTRTDSYS AGQSVEILDG VELGEPAHKK TGTTVPESIH SFIGDGLVK PEALNKKAIQ IINRVRDKLT GRDFSHDDTL DVPTQVELLI KQATSHENLC QCYIGWCPFW UniProtKB: Serine/threonine-protein kinase mTOR |
-Macromolecule #2: Target of rapamycin complex subunit LST8
| Macromolecule | Name: Target of rapamycin complex subunit LST8 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.91009 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNTSPGTVGS DPVILATAGY DHTVRFWQAH SGICTRTVQH QDSQVNALEV TPDRSMIAAA GYQHIRMYDL NSNNPNPIIS YDGVNKNIA SVGFHEDGRW MYTGGEDCTA RIWDLRSRNL QCQRIFQVNA PINCVCLHPN QAELIVGDQS GAIHIWDLKT D HNEQLIPE ...String: MNTSPGTVGS DPVILATAGY DHTVRFWQAH SGICTRTVQH QDSQVNALEV TPDRSMIAAA GYQHIRMYDL NSNNPNPIIS YDGVNKNIA SVGFHEDGRW MYTGGEDCTA RIWDLRSRNL QCQRIFQVNA PINCVCLHPN QAELIVGDQS GAIHIWDLKT D HNEQLIPE PEVSITSAHI DPDASYMAAV NSTGNCYVWN LTGGIGDEVT QLIPKTKIPA HTRYALQCRF SPDSTLLATC SA DQTCKIW RTSNFSLMTE LSIKSGNPGE SSRGWMWGCA FSGDSQYIVT ASSDNLARLW CVETGEIKRE YGGHQKAVVC LAF NDSVLG UniProtKB: Target of rapamycin complex subunit LST8 |
-Macromolecule #3: Regulatory-associated protein of mTOR
| Macromolecule | Name: Regulatory-associated protein of mTOR / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 152.764656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHHH EQKLISEEDL DYKDDDDKME SEMLQSPLLG LGEEDEADLT DWNLPLAFMK KRHCEKIEGS KSLAQSWRMK DRMKTVSVA LVLCLNVGVD PPDVVKTTPC ARLECWIDPL SMGPQKALET IGANLQKQYE NWQPRARYKQ SLDPTVDEVK K LCTSLRRN ...String: HHHHHHHHHH EQKLISEEDL DYKDDDDKME SEMLQSPLLG LGEEDEADLT DWNLPLAFMK KRHCEKIEGS KSLAQSWRMK DRMKTVSVA LVLCLNVGVD PPDVVKTTPC ARLECWIDPL SMGPQKALET IGANLQKQYE NWQPRARYKQ SLDPTVDEVK K LCTSLRRN AKEERVLFHY NGHGVPRPTV NGEVWVFNKN YTQYIPLSIY DLQTWMGSPS IFVYDCSNAG LIVKSFKQFA LQ REQELEV AAINPNHPLA QMPLPPSMKN CIQLAACEAT ELLPMIPDLP ADLFTSCLTT PIKIALRWFC MQKCVSLVPG VTL DLIEKI PGRLNDRRTP LGELNWIFTA ITDTIAWNVL PRDLFQKLFR QDLLVASLFR NFLLAERIMR SYNCTPVSSP RLPP TYMHA MWQAWDLAVD ICLSQLPTII EEGTAFRHSP FFAEQLTAFQ VWLTMGVENR NPPEQLPIVL QVLLSQVHRL RALDL LGRF LDLGPWAVSL ALSVGIFPYV LKLLQSSARE LRPLLVFIWA KILAVDSSCQ ADLVKDNGHK YFLSVLADPY MPAEHR TMT AFILAVIVNS YHTGQEACLQ GNLIAICLEQ LNDPHPLLRQ WVAICLGRIW QNFDSARWCG VRDSAHEKLY SLLSDPI PE VRCAAVFALG TFVGNSAERT DHSTTIDHNV AMMLAQLVSD GSPMVRKELV VALSHLVVQY ESNFCTVALQ FIEEEKNY A LPSPATTEGG SLTPVRDSPC TPRLRSVSSY GNIRAVATAR SLNKSLQNLS LTEESGGAVA FSPGNLSTSS SASSTLGSP ENEEHILSFE TIDKMRRASS YSSLNSLIGV SFNSVYTQIW RVLLHLAADP YPEVSDVAMK VLNSIAYKAT VNARPQRVLD TSSLTQSAP ASPTNKGVHI HQAGGSPPAS STSSSSLTND VAKQPVSRDL PSGRPGTTGP AGAQYTPHSH QFPRTRKMFD K GPEQTADD ADDAAGHKSF ISATVQTGFC DWSARYFAQP VMKIPEEHDL ESQIRKEREW RFLRNSRVRR QAQQVIQKGI TR LDDQIFL NRNPGVPSVV KFHPFTPCIA VADKDSICFW DWEKGEKLDY FHNGNPRYTR VTAMEYLNGQ DCSLLLTATD DGA IRVWKN FADLEKNPEM VTAWQGLSDM LPTTRGAGMV VDWEQETGLL MSSGDVRIVR IWDTDREMKV QDIPTGADSC VTSL SCDSH RSLIVAGLGD GSIRVYDRRM ALSECRVMTY REHTAWVVKA SLQKRPDGHI VSVSVNGDVR IFDPRMPESV NVLQI VKGL TALDIHPQAD LIACGSVNQF TAIYNSSGEL INNIKYYDGF MGQRVGAISC LAFHPHWPHL AVGSNDYYIS VYSVEK RVR UniProtKB: Regulatory-associated protein of mTOR |
-Macromolecule #4: Lateral signaling target protein 2 homolog
| Macromolecule | Name: Lateral signaling target protein 2 homolog / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 96.604477 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMNRFRKWLY KPKRSDPQLL ARFYYADEEL NQVAAELDSL DGRKDPQRCT LLVSQFRSCQ DNVLNIINQI MDECIPQDRA PRDFCVKFP EEIRHDNLAG QLWFGAECLA AGSIIMNREL ESMAMRPLAK ELTRSLEDVR GALRDQALRD LNTYTEKMRE A LRHFDVLF ...String: MMNRFRKWLY KPKRSDPQLL ARFYYADEEL NQVAAELDSL DGRKDPQRCT LLVSQFRSCQ DNVLNIINQI MDECIPQDRA PRDFCVKFP EEIRHDNLAG QLWFGAECLA AGSIIMNREL ESMAMRPLAK ELTRSLEDVR GALRDQALRD LNTYTEKMRE A LRHFDVLF AEFELSYVSA MVPVKSPREY YVQQEVIVLF CETVERALDF GYLTQDMIDD YEPALMFSIP RLAIVCGLVV YA DGPLNLD RKVEDMSELF RPFHTLLRKI RDLLQTLTEE ELHTLERNLC ISQDVEFPIR ADVQGPAALA PALSAPLPPE GPL SAKAKD PDAELACSMQ YDDQELEQLS RMVHRAGDEM SSLLSPPIAC QSPAHRPGAE GSPGGEASPG RPRLRSGSDE EERV FFMDD VEGTAEALAR PESPAGPFGW AGSTWADPQE KGQGGPGGAA GISLPASEKE EDLSNNNLEA EGTDGASLAG TSSCS CLDS RLHLDGWEVG ADDAETAEMI AHRTGGMKLS ATVIFNPKSP TSLDSAVATQ EAASEPVAEG MDGGPHKLST GATNCL LHS CVCCGSCGDS REDVVERLRE KCSPGGVIGA SYAAGLAKAS DRAPERQEEA PPPSEDASNG REPKAPTSDK CLPHTSG SQ VDTASGLQGE AGVAGQQEPE ARELHAGSPS AHEAPQALSG SSSSTAGSCS SDKMGPEAAP AATHAAPQAT REKIRSRF H GSHDLIHRLF VCISGVADQL QTNYASDLRS ILKTLFEVMA TKPETDDKEK LRKVTQTLRS AALEDCALCQ ETLSSSELA AKTRDGDFED PPEWVPDEAC GFCTACKAPF TVIRRKHHCR SCGKIFCSRC SSHSAPLPRY GQVKPVRVCT HCYMFHVTPF YSDKAGL UniProtKB: Lateral signaling target protein 2 homolog |
-Macromolecule #5: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4719 / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)