[English] 日本語
 Yorodumi
Yorodumi- EMDB-50181: cryo-EM structure of LST2 TOS peptide bound to human mTOR complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of LST2 TOS peptide bound to human mTOR complex 1, focused on RAPTOR | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MTOR / MTORC1 / LST2 / ZFYVE28 / EGFR / TOS / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of epidermal growth factor-activated receptor activity / positive regulation of pentose-phosphate shunt / TORC1 complex / positive regulation of odontoblast differentiation / MTOR signalling / cellular response to L-leucine / Energy dependent regulation of mTOR by LKB1-AMPK / Amino acids regulate mTORC1 / phosphatidylinositol-3-phosphate binding / serine/threonine protein kinase complex ...negative regulation of epidermal growth factor-activated receptor activity / positive regulation of pentose-phosphate shunt / TORC1 complex / positive regulation of odontoblast differentiation / MTOR signalling / cellular response to L-leucine / Energy dependent regulation of mTOR by LKB1-AMPK / Amino acids regulate mTORC1 / phosphatidylinositol-3-phosphate binding / serine/threonine protein kinase complex / positive regulation of osteoclast differentiation / cellular response to osmotic stress / positive regulation of transcription by RNA polymerase III / TORC1 signaling / Macroautophagy / regulation of cell size / negative regulation of epidermal growth factor receptor signaling pathway / TOR signaling / mTORC1-mediated signalling / HSF1-dependent transactivation / social behavior / positive regulation of TOR signaling / enzyme-substrate adaptor activity / positive regulation of G1/S transition of mitotic cell cycle / protein serine/threonine kinase inhibitor activity / protein kinase activator activity / positive regulation of lipid biosynthetic process / 14-3-3 protein binding / positive regulation of endothelial cell proliferation / cellular response to nutrient levels / negative regulation of autophagy / cellular response to starvation / positive regulation of glycolytic process / Regulation of PTEN gene transcription / TP53 Regulates Metabolic Genes / regulation of cell growth / cellular response to amino acid stimulus / cellular response to glucose stimulus / regulation of autophagy / small GTPase binding / cytoplasmic stress granule / positive regulation of cell growth / early endosome membrane / cellular response to hypoxia / protein-macromolecule adaptor activity / lysosome / response to xenobiotic stimulus / lysosomal membrane / neuronal cell body / DNA damage response / dendrite / protein kinase binding / protein-containing complex binding / zinc ion binding / nucleoplasm / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | |||||||||
 Authors Authors | Craigie LM / Maier T | |||||||||
| Funding support |  Switzerland, European Union, 2 items Switzerland, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: mTORC1 phosphorylates and stabilizes LST2 to negatively regulate EGFR. Authors: Stefania Battaglioni / Louise-Marie Craigie / Sofia Filippini / Timm Maier / Michael N Hall /  Abstract: TORC1 (target of rapamycin complex 1) is a highly conserved protein kinase that plays a central role in regulating cell growth. Given the role of mammalian TORC1 (mTORC1) in metabolism and disease, ...TORC1 (target of rapamycin complex 1) is a highly conserved protein kinase that plays a central role in regulating cell growth. Given the role of mammalian TORC1 (mTORC1) in metabolism and disease, understanding mTORC1 downstream signaling and feedback loops is important. mTORC1 recognizes some of its substrates via a five amino acid binding sequence called the TOR signaling (TOS) motif. mTORC1 binding to a TOS motif facilitates phosphorylation of a distinct, distal site. Here, we show that LST2, also known as ZFYVE28, contains a TOS motif (amino acids 401 to 405) and is directly phosphorylated by mTORC1 at serine 670 (S670). mTORC1-mediated S670 phosphorylation promotes LST2 monoubiquitination on lysine 87 (K87). Monoubiquitinated LST2 is stable and displays a broad reticular distribution. When mTORC1 is inactive, unphosphorylated LST2 is degraded by the proteasome. The absence of LST2 enhances EGFR (epidermal growth factor receptor) signaling. We propose that mTORC1 negatively feeds back on its upstream receptor EGFR via LST2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50181.map.gz emd_50181.map.gz | 483.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50181-v30.xml emd-50181-v30.xml emd-50181.xml emd-50181.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
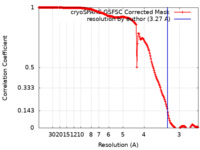
| FSC (resolution estimation) |  emd_50181_fsc.xml emd_50181_fsc.xml | 16.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_50181.png emd_50181.png | 81.9 KB | ||
| Masks |  emd_50181_msk_1.map emd_50181_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50181.cif.gz emd-50181.cif.gz | 6.7 KB | ||
| Others |  emd_50181_half_map_1.map.gz emd_50181_half_map_1.map.gz emd_50181_half_map_2.map.gz emd_50181_half_map_2.map.gz | 475.8 MB 475.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50181 http://ftp.pdbj.org/pub/emdb/structures/EMD-50181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50181 | HTTPS FTP |
-Related structure data
| Related structure data |  9f42MC  9f43C  9f44C  9f45C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50181.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50181.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||
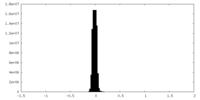
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50181_msk_1.map emd_50181_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
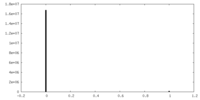
| Density Histograms |
-Half map: #2
| File | emd_50181_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
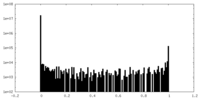
| Density Histograms |
-Half map: #1
| File | emd_50181_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
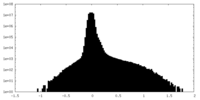
| Density Histograms |
- Sample components
Sample components
-Entire : mTORC1 in complex with synthesised LST2 TOS peptide
| Entire | Name: mTORC1 in complex with synthesised LST2 TOS peptide |
|---|---|
| Components |
|
-Supramolecule #1: mTORC1 in complex with synthesised LST2 TOS peptide
| Supramolecule | Name: mTORC1 in complex with synthesised LST2 TOS peptide / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Regulatory-associated protein of mTOR
| Macromolecule | Name: Regulatory-associated protein of mTOR / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 152.764656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHHH EQKLISEEDL DYKDDDDKME SEMLQSPLLG LGEEDEADLT DWNLPLAFMK KRHCEKIEGS KSLAQSWRMK DRMKTVSVA LVLCLNVGVD PPDVVKTTPC ARLECWIDPL SMGPQKALET IGANLQKQYE NWQPRARYKQ SLDPTVDEVK K LCTSLRRN ...String: HHHHHHHHHH EQKLISEEDL DYKDDDDKME SEMLQSPLLG LGEEDEADLT DWNLPLAFMK KRHCEKIEGS KSLAQSWRMK DRMKTVSVA LVLCLNVGVD PPDVVKTTPC ARLECWIDPL SMGPQKALET IGANLQKQYE NWQPRARYKQ SLDPTVDEVK K LCTSLRRN AKEERVLFHY NGHGVPRPTV NGEVWVFNKN YTQYIPLSIY DLQTWMGSPS IFVYDCSNAG LIVKSFKQFA LQ REQELEV AAINPNHPLA QMPLPPSMKN CIQLAACEAT ELLPMIPDLP ADLFTSCLTT PIKIALRWFC MQKCVSLVPG VTL DLIEKI PGRLNDRRTP LGELNWIFTA ITDTIAWNVL PRDLFQKLFR QDLLVASLFR NFLLAERIMR SYNCTPVSSP RLPP TYMHA MWQAWDLAVD ICLSQLPTII EEGTAFRHSP FFAEQLTAFQ VWLTMGVENR NPPEQLPIVL QVLLSQVHRL RALDL LGRF LDLGPWAVSL ALSVGIFPYV LKLLQSSARE LRPLLVFIWA KILAVDSSCQ ADLVKDNGHK YFLSVLADPY MPAEHR TMT AFILAVIVNS YHTGQEACLQ GNLIAICLEQ LNDPHPLLRQ WVAICLGRIW QNFDSARWCG VRDSAHEKLY SLLSDPI PE VRCAAVFALG TFVGNSAERT DHSTTIDHNV AMMLAQLVSD GSPMVRKELV VALSHLVVQY ESNFCTVALQ FIEEEKNY A LPSPATTEGG SLTPVRDSPC TPRLRSVSSY GNIRAVATAR SLNKSLQNLS LTEESGGAVA FSPGNLSTSS SASSTLGSP ENEEHILSFE TIDKMRRASS YSSLNSLIGV SFNSVYTQIW RVLLHLAADP YPEVSDVAMK VLNSIAYKAT VNARPQRVLD TSSLTQSAP ASPTNKGVHI HQAGGSPPAS STSSSSLTND VAKQPVSRDL PSGRPGTTGP AGAQYTPHSH QFPRTRKMFD K GPEQTADD ADDAAGHKSF ISATVQTGFC DWSARYFAQP VMKIPEEHDL ESQIRKEREW RFLRNSRVRR QAQQVIQKGI TR LDDQIFL NRNPGVPSVV KFHPFTPCIA VADKDSICFW DWEKGEKLDY FHNGNPRYTR VTAMEYLNGQ DCSLLLTATD DGA IRVWKN FADLEKNPEM VTAWQGLSDM LPTTRGAGMV VDWEQETGLL MSSGDVRIVR IWDTDREMKV QDIPTGADSC VTSL SCDSH RSLIVAGLGD GSIRVYDRRM ALSECRVMTY REHTAWVVKA SLQKRPDGHI VSVSVNGDVR IFDPRMPESV NVLQI VKGL TALDIHPQAD LIACGSVNQF TAIYNSSGEL INNIKYYDGF MGQRVGAISC LAFHPHWPHL AVGSNDYYIS VYSVEK RVR UniProtKB: Regulatory-associated protein of mTOR |
-Macromolecule #2: Lateral signaling target protein 2 homolog
| Macromolecule | Name: Lateral signaling target protein 2 homolog / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.660733 KDa |
| Sequence | String: DEEERVFFMD DVE UniProtKB: Lateral signaling target protein 2 homolog |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4470 / Average electron dose: 48.43 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)