+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
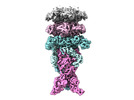
| Title | Cryo-EM of tail-tip of bacteriophage Chi | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Flagellotropic bacteriophage / Siphophage / Tail-tip / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Chivirus chi Chivirus chi | |||||||||
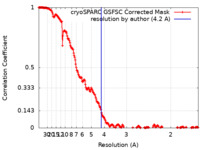
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Sonani RR / Esteves NC / Scharf BE / Egelman EH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Cryo-EM structure of flagellotropic bacteriophage Chi. Authors: Ravi R Sonani / Nathaniel C Esteves / Birgit E Scharf / Edward H Egelman /  Abstract: The flagellotropic bacteriophage χ (Chi) infects bacteria via the flagellar filament. Despite years of study, its structural architecture remains partly characterized. Through cryo-EM, we unveil ...The flagellotropic bacteriophage χ (Chi) infects bacteria via the flagellar filament. Despite years of study, its structural architecture remains partly characterized. Through cryo-EM, we unveil χ's nearly complete structure, encompassing capsid, neck, tail, and tail tip. While the capsid and tail resemble phage YSD1, the neck and tail tip reveal new proteins and their arrangement. The neck shows a unique conformation of the tail tube protein, forming a socket-like structure for attachment to the neck. The tail tip comprises four proteins, including distal tail protein (DTP), two baseplate hub proteins (BH1P and BH2P), and tail tip assembly protein (TAP) exhibiting minimal organization compared to other siphophages. Deviating from the consensus in other siphophages, DTP in χ forms a trimeric assembly, reducing tail symmetry from 6-fold to 3-fold at the tip. These findings illuminate the previously unexplored structural organization of χ's neck and tail tip. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43281.map.gz emd_43281.map.gz | 777.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43281-v30.xml emd-43281-v30.xml emd-43281.xml emd-43281.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43281_fsc.xml emd_43281_fsc.xml | 20 KB | Display |  FSC data file FSC data file |
| Images |  emd_43281.png emd_43281.png | 64.7 KB | ||
| Masks |  emd_43281_msk_1.map emd_43281_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43281.cif.gz emd-43281.cif.gz | 8 KB | ||
| Others |  emd_43281_additional_1.map.gz emd_43281_additional_1.map.gz emd_43281_half_map_1.map.gz emd_43281_half_map_1.map.gz emd_43281_half_map_2.map.gz emd_43281_half_map_2.map.gz | 763.3 MB 764.7 MB 764.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43281 http://ftp.pdbj.org/pub/emdb/structures/EMD-43281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43281 | HTTPS FTP |
-Validation report
| Summary document |  emd_43281_validation.pdf.gz emd_43281_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43281_full_validation.pdf.gz emd_43281_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_43281_validation.xml.gz emd_43281_validation.xml.gz | 28.5 KB | Display | |
| Data in CIF |  emd_43281_validation.cif.gz emd_43281_validation.cif.gz | 37.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43281 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43281 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43281 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43281 | HTTPS FTP |
-Related structure data
| Related structure data |  8vjhMC  8vhxC  8vjaC  8vjiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43281.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43281.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
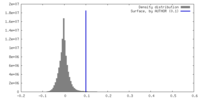
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43281_msk_1.map emd_43281_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
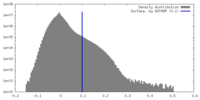
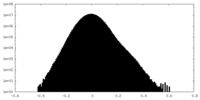
| Density Histograms |
-Additional map: #1
| File | emd_43281_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
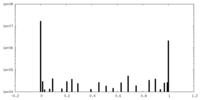
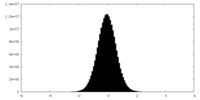
| Density Histograms |
-Half map: #1
| File | emd_43281_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
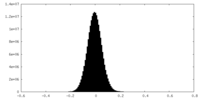
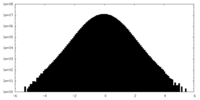
| Density Histograms |
-Half map: #2
| File | emd_43281_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
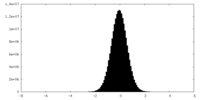
| Density Histograms |
- Sample components
Sample components
-Entire : Chivirus chi
| Entire | Name:  Chivirus chi Chivirus chi |
|---|---|
| Components |
|
-Supramolecule #1: Chivirus chi
| Supramolecule | Name: Chivirus chi / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 / NCBI-ID: 1541887 / Sci species name: Chivirus chi / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Minor tail protein
| Macromolecule | Name: Minor tail protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chivirus chi Chivirus chi |
| Molecular weight | Theoretical: 143.342531 KDa |
| Sequence | String: MGSKSKKVTV GYKYYMGLFM GLFRGPVNEI TEIRVGDRTA WTGSITGNTT IQINKESLFG GTKAEGGIDG PLEVYMGAPT QTVSQKLKN MLGGRQPEFR GVVTAYFDGL ICAMNPYPKQ WKFKARRSTA GWTGGVWYPE KCLVKMQGYD GQGNQHEIHA M NPAHILYE ...String: MGSKSKKVTV GYKYYMGLFM GLFRGPVNEI TEIRVGDRTA WTGSITGNTT IQINKESLFG GTKAEGGIDG PLEVYMGAPT QTVSQKLKN MLGGRQPEFR GVVTAYFDGL ICAMNPYPKQ WKFKARRSTA GWTGGVWYPE KCLVKMQGYD GQGNQHEIHA M NPAHILYE CQSNYEWGRG LSRTLIDDTT FRLAADTLFN ENFGLCIRWN RQDTLESFMQ LILDHIGGAM YVSKVTGKLS LR LIRKDYD FDTLPIFDTD SGLLSIQEAT NASPANLVNE VVVTYHNPVM DEDQQVRSHN LAQIQNQGCL NSNTIEYLGI PTG KLAMQV AQRDLRAAST NVRRFTVICD RRAWNVQPGD VLKVRDPKQR GLTEVVIRVG TVEDGTLPDG KIKVVALQDQ FAFQ LNTFN QVEPPAGYEP NLTPAIARRI VYEMPYVDLV QQLPDGELNA VGPNDAFINS QAEKPTDMSA AYDMGIMAEG ESGFD VRGN GDFGAFGSLA SDIDYLSTQI VLSEMTDMWD DVQPGFVARI AKPVLDGQRT QMEIAEEFVR IDAINGNVIT VARGVL DTI PFRHQKSEML WVTTYDGGTD WQKYAGNESI DIKILPWTLG GGRFPIEDAP IDHLDMDFRQ IRPYPPGNVQ HYLASTS TL QRWYVPSALT YTANAGETPD TYTLTWAHRD RVLQGDKPVP HIDGDIGPEP GTSYTIRVYN QEGTLVRTES GINGTTWN W PYATAAADMN VEASLFDPVL ATLRLTSARN GRESWEYYEM KVSVYKKPPQ FVYDASLMQF AAQPYNATSD PNPPTPPMD GPYVSSLMQF AAQPDGSEDF GPAESMNGPN VALLPHQVTQ TSSIITPLDT LLYETPYIQL SRDGRDLNTS KVSAYVARSS DRTVDSYTL FTKHETDANY TSSGTQPWTP WGMSTVGLGF FTDEVTIDPT SDKDGVPVAP AKVGDLLLID QELVSITAIN G RTYKIGRG VADTIPAQHY SSRPVWLISV GFGYSDMAFG DNEKAMVIIR PDTYGTDIPL SKLYPLQLQM QYRPKRPYPP GL MMIGGQP FFNTASGLAD DFNPYDNLKA KDVVCTYAHR NRVSQSTTAR DHFAVGITPE PGVKYRVRIG YAYWSQSGSG FSL LSQFET EDAGFIMRAA DLERWGRQAG YAQDAGGYST LNVTVNAIRD GMLNWQGYTM TVRVPSFPLP PGQKPGGGDG PWNP PGGGN TGGTTPPTEP PDRPDPSEPG TTDPSEPTKP DEPEEPQPPV DPEDPDTDPP EPPEPPVDPT NVPGWSLSWD HGWAM TLPD FRYVPPADEE PE UniProtKB: Minor tail protein |
-Macromolecule #2: Distal tail protein
| Macromolecule | Name: Distal tail protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chivirus chi Chivirus chi |
| Molecular weight | Theoretical: 63.032605 KDa |
| Sequence | String: MALSGLRPAR EQFNQAIYDA LNGAYQLYVQ DAFGQAAANQ TYYMAKWMKL KAGTYTAVMY VDDSGTLSID HAVVATAAIG TNPNSGEFT VAADGVYRFD CIYSNVPADT PAYMAYQLIR DGQTVEVSRA NDFIADIVPI PDKALGPKPP YSDDVRLTYP V FLPLPNWK ...String: MALSGLRPAR EQFNQAIYDA LNGAYQLYVQ DAFGQAAANQ TYYMAKWMKL KAGTYTAVMY VDDSGTLSID HAVVATAAIG TNPNSGEFT VAADGVYRFD CIYSNVPADT PAYMAYQLIR DGQTVEVSRA NDFIADIVPI PDKALGPKPP YSDDVRLTYP V FLPLPNWK DGVTERIEWQ TDVMISESGA EQRRPIRLHP RRSFEATFLR WEENRTLLDT TIAGVGQSPL LLPLWHDMTA TE NNAPAGS VDIFGQFRVK DFNVGDVVMF NRGTTWDYET NIIAGLDIDA GHMTLTFGLQ SDTPKGTRLY PVRVAQIREA MNG QQMTDS VSQTQVRFFC TENYDLTPSW SDFPIYTRTG LHIFVLPEDW GSSNEITSDR LTYNFDNQSG PVVVVDPGGQ NYGT VKKSY TIKGRTADRQ FRQILFALRG RTKTFHLPLD TNDFILSRDI NPADGALVVR RCGYTQYIGG TQETKRDIMV ELYDG TRIP TTIISSRIVG DEEWLFLSQS IPATSRNDVR RIGYIPVARL DVDGIEIKRL TDSAGVSQVS LTFKFFDDRR IATPLP LS UniProtKB: Distal tail protein |
-Macromolecule #3: Baseplate hub 1 protein, gp27
| Macromolecule | Name: Baseplate hub 1 protein, gp27 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chivirus chi Chivirus chi |
| Molecular weight | Theoretical: 29.348869 KDa |
| Sequence | String: MSYNIIETSN DNGRPVFMYE FRLLDKYWRY TSADAKVSAL GSIWEPMGVS DDGIKQTGEA KTDALNLTLP NSNPVVGLFI GTPPGSPVT LTIRRMHLDD NDPVVCYVGT VDSINQGENP TVATVTCSTL SATMDRNGLR LSWSRGCPHA LYDGQCRVNK E AFRVDATI ...String: MSYNIIETSN DNGRPVFMYE FRLLDKYWRY TSADAKVSAL GSIWEPMGVS DDGIKQTGEA KTDALNLTLP NSNPVVGLFI GTPPGSPVT LTIRRMHLDD NDPVVCYVGT VDSINQGENP TVATVTCSTL SATMDRNGLR LSWSRGCPHA LYDGQCRVNK E AFRVDATI LTVGAGTVTA AAYATRPDGY FAGGFIEWID PVYGVERRGI ETHTGNTITI FGTVDGLAGG YILKTYPGCP RT SAACDTI FNNLANFGGI PSLPDRSPFD GNPIF UniProtKB: Bacteriophage phiJL001 Gp84 C-terminal domain-containing protein |
-Macromolecule #4: Tail Tube
| Macromolecule | Name: Tail Tube / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chivirus chi Chivirus chi |
| Molecular weight | Theoretical: 40.375363 KDa |
| Sequence | String: MNDNYQNNYV VGRGTVYFDR FQDGTNRKTG EMYFGNTPEF TINTDSETLD HYSSDHGMRV MDASVLLEAS QGGTFTCDNI NADNLALWF LGEVSNTTQT QQTDAKEVFN PIMRGRYYQL GTTDDNPTGV RGVTNFQMVK ADASIAISVG SGDITSIVGA T VVNPAGNY ...String: MNDNYQNNYV VGRGTVYFDR FQDGTNRKTG EMYFGNTPEF TINTDSETLD HYSSDHGMRV MDASVLLEAS QGGTFTCDNI NADNLALWF LGEVSNTTQT QQTDAKEVFN PIMRGRYYQL GTTDDNPTGV RGVTNFQMVK ADASIAISVG SGDITSIVGA T VVNPAGNY EIDLEAGRIY IEPDSTDLSG NVQIAVQYDV DAQKRTLVIG KSNMVYGALR MISDNPVGLN KNYYFPKVSI AP DGDYALK GDDWQVMSFT FKAMQLNNIT QRVYIDIVEA AAAVDPTAQR TIEITPASTT ATTGGAGVVC TVTVRDGTGT AVQ GDAVTF TTVAGATVTP NSATTGATGT ATTTVNRAAA GTATVTATLA NGKAATTGTI TFSAP UniProtKB: Major tail protein |
-Macromolecule #5: Tape measure
| Macromolecule | Name: Tape measure / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chivirus chi Chivirus chi |
| Molecular weight | Theoretical: 153.960453 KDa |
| Sequence | String: MANSKDVELR IRARDYSQKP LKAVTSAIEQ MAKAQDEQRK AAERGEISTR DLEAAYKKLE SAGSQLLKLN SLIEVFKRQN QAMTEATAK TEGLRQKQAD LQKTYDSTEK VTQKQERALA RVTRQVEAAT RAEANQAERV NRATRDLERY GIETSKVGAA Q AGIVTSVA ...String: MANSKDVELR IRARDYSQKP LKAVTSAIEQ MAKAQDEQRK AAERGEISTR DLEAAYKKLE SAGSQLLKLN SLIEVFKRQN QAMTEATAK TEGLRQKQAD LQKTYDSTEK VTQKQERALA RVTRQVEAAT RAEANQAERV NRATRDLERY GIETSKVGAA Q AGIVTSVA QVNRVLERQD EIISTSAAAA AQAKVIRGLQ QQADQAMATA RGYQTLGRVV QQATGQLGPL GTQIQTIVSP AE AARRTLS GLENQVHGVT AELARNSKEV ENVAQKVRML NEANKTVSAL AQQIDLYRQQ VATLRNARTE YQQARQDVIK LAQ QMRTAT TDTGALGIQM QAAQQRLSAA ATAMRNTATA ARSTQAALRA AQVDTRNLSD AEARLISTSQ QSAAALNTLS TATN RNSQA ARDGSKAWSL FRDEGRTTLS FLQRIRGEVL ALTTTYVGFQ GAISLAGGAI DAYKNRQQAM VKIANVVGNS QAAIN KEWE YMVGLANTLG IDITTLSQSY TKFAVSAKAV GLSLQDSKFI FESVAKAGRV FHLSQDDMEG VFRALEQMLS KGQVYA EEL RGQLGERLPA AFALFAKGMD MTTAELMKAM ENGEVTGEAV INFAREQAKA IDAQLATAQK GVDAMEARAR NAMNAFQ LA LADAGFIEAY VQMLNKITDF LNSEDGRAAA VKLGEAFGML ADAVTWCIEN VDTLITALSI LAGLKVVQFI GGMISGLK N LLPLFNTLSK IGAGIITVLE GVAARMIAAE GAIGLLGVAL KGLTRLIPIV GWALLAYDIG AIMYDQSQTF REAVNAVIR DFKNLGNQLA AVVDTIPALL YDLAVSVLRP ITTMFADTTQ AIIKWIADVL KLIPGVGQGL SEWATSIGDD LTKEQRGFLE STGRIWDNV NKQWVKLNDD MVAKNADATD KIRGQVNQLA ADMAAITKGE GFQFTQDPGT GVTKRSREIA GLTKELNKME E AAKKADVA SRKAEQRKNL SGRLAIIDEE FAPQYARAKS IGGSEGDALT KRLDAVVAAR KKAETTLFNS QQRTTGGIKK QE NALQALI NKYNELNAAV GVKEVKIDPN ATFDDRLAAK LAAVNTQYDQ LIAKSKKLGT GGENLAGQFE DLRKRNLEYA TTQ AKLEEL KRIQDQLNAQ QETRKNLLDE INAKRQAGII SEDEAVSQTV ALYQNMNAGI ASSAEQLDVF AQKIKDTMSP EEFS RIMAQ IAAVKAGLVD VTGTFTTMDT TVVQGVLDGM STALSSIVDE MALVVAGSQS IGDAFSNLGV AVARFFADFL QKIAM AILQ QMALNALASM GGGIGSAAVA LGGTVAKHNG GTVGSKTTGG TQMKGGISPA MFANAPRFHD GGLPGLRSDE VPTILQ KGE QVLSKDDPNN VLNQSRGGGQ TAQSPQGLRF VLVDDRSKVP EAMNTPEGEK AVMQILQRNV PTLKNLVG UniProtKB: Tape measure |
-Macromolecule #6: Tail assembly protein 1
| Macromolecule | Name: Tail assembly protein 1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chivirus chi Chivirus chi |
| Molecular weight | Theoretical: 8.278664 KDa |
| Sequence | String: MWWALAMLVA SVLINAALAP KPASAKPATI QDFDIPQVKE GTPQSVVFGE VWTADWQVLG FGNFRTKAVK AKQAKK UniProtKB: Tail assembly protein 1 |
-Macromolecule #7: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | Tail-tip of bacteriophage Chi |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)