+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of capsid of bacteriophage Chi | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Flagellotropic bacteriophage / Siphophage / Capsid / VIRUS | |||||||||
| Function / homology | Head decoration protein D / Bacteriophage lambda head decoration protein D / Major capsid protein GpE / Phage major capsid protein E / viral capsid / host cell cytoplasm / Decorator protein D / Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Chivirus chi Chivirus chi | |||||||||
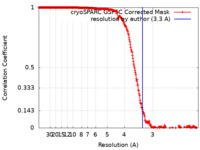
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Sonani RR / Esteves NC / Scharf BE / Egelman EH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Cryo-EM structure of flagellotropic bacteriophage Chi. Authors: Ravi R Sonani / Nathaniel C Esteves / Birgit E Scharf / Edward H Egelman /  Abstract: The flagellotropic bacteriophage χ (Chi) infects bacteria via the flagellar filament. Despite years of study, its structural architecture remains partly characterized. Through cryo-EM, we unveil ...The flagellotropic bacteriophage χ (Chi) infects bacteria via the flagellar filament. Despite years of study, its structural architecture remains partly characterized. Through cryo-EM, we unveil χ's nearly complete structure, encompassing capsid, neck, tail, and tail tip. While the capsid and tail resemble phage YSD1, the neck and tail tip reveal new proteins and their arrangement. The neck shows a unique conformation of the tail tube protein, forming a socket-like structure for attachment to the neck. The tail tip comprises four proteins, including distal tail protein (DTP), two baseplate hub proteins (BH1P and BH2P), and tail tip assembly protein (TAP) exhibiting minimal organization compared to other siphophages. Deviating from the consensus in other siphophages, DTP in χ forms a trimeric assembly, reducing tail symmetry from 6-fold to 3-fold at the tip. These findings illuminate the previously unexplored structural organization of χ's neck and tail tip. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43282.map.gz emd_43282.map.gz | 1.6 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43282-v30.xml emd-43282-v30.xml emd-43282.xml emd-43282.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43282_fsc.xml emd_43282_fsc.xml | 25.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_43282.png emd_43282.png | 85.2 KB | ||
| Filedesc metadata |  emd-43282.cif.gz emd-43282.cif.gz | 5.2 KB | ||
| Others |  emd_43282_half_map_1.map.gz emd_43282_half_map_1.map.gz emd_43282_half_map_2.map.gz emd_43282_half_map_2.map.gz | 1.6 GB 1.6 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43282 http://ftp.pdbj.org/pub/emdb/structures/EMD-43282 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43282 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43282 | HTTPS FTP |
-Related structure data
| Related structure data |  8vjiMC  8vhxC  8vjaC  8vjhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43282.map.gz / Format: CCP4 / Size: 1.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43282.map.gz / Format: CCP4 / Size: 1.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0677 Å | ||||||||||||||||||||||||||||||||||||
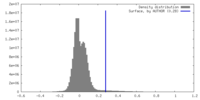
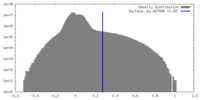
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_43282_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
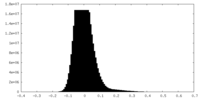
| Density Histograms |
-Half map: #1
| File | emd_43282_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
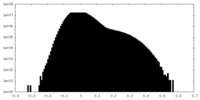
| Density Histograms |
- Sample components
Sample components
-Entire : Chivirus chi
| Entire | Name:  Chivirus chi Chivirus chi |
|---|---|
| Components |
|
-Supramolecule #1: Chivirus chi
| Supramolecule | Name: Chivirus chi / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1541887 / Sci species name: Chivirus chi / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chivirus chi Chivirus chi |
| Molecular weight | Theoretical: 39.959625 KDa |
| Sequence | String: MAGLYTTYQL LEVQRKLKTL PAFFLQWFPR QINFQEDMIA FDKVIQDVTR VAPFVAPNVQ GRVIKESGYN TKTFKPAYVK PKHVIDPNM IIPRQPGEAL GTGTLSIAQR RDRVIAYLLM KHRAMHENTW EWMAAQAAQY GYVDVQGQDY PLVRVDFGRD A ALTMTTDW ...String: MAGLYTTYQL LEVQRKLKTL PAFFLQWFPR QINFQEDMIA FDKVIQDVTR VAPFVAPNVQ GRVIKESGYN TKTFKPAYVK PKHVIDPNM IIPRQPGEAL GTGTLSIAQR RDRVIAYLLM KHRAMHENTW EWMAAQAAQY GYVDVQGQDY PLVRVDFGRD A ALTMTTDW TAAGVTLMDM IADLRDGQRL VSDKSMSGTV IRDYIFGGDA WDQFVKVGGK ELWGKDGLMD STIRGSETNV TR LWDDVEG VQYMGELVGA NGAGRMRIWV NTQKYRDQND QEQFLMKQKA VMGISSAIEG VRCFGAILDK GAGYQALDYF PKM WDQEDP SVEYLMSQGA PLMVPADPNA SFLLTVMS UniProtKB: Major capsid protein |
-Macromolecule #2: Decorator protein D
| Macromolecule | Name: Decorator protein D / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chivirus chi Chivirus chi |
| Molecular weight | Theoretical: 14.401047 KDa |
| Sequence | String: MNLLTMMAAT SLPNYLAGNG DLGSWEPTQI FAGEADIVTE GGAAGADIEI YQVIAKNAAG AMVPHDPTAT TGTSPDEVPA PQSVAIGIA AQPAKSGQNV PYYIGGVFNH AALGWHASLD TLAKRQAVFD RTNIHIGNLY UniProtKB: Decorator protein D |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | Capsid of bacteriophage Chi |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)