[English] 日本語
 Yorodumi
Yorodumi- EMDB-41584: Actin 1 from T. gondii in filaments bound to MgADP and jasplakinolide -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Actin 1 from T. gondii in filaments bound to MgADP and jasplakinolide | |||||||||
 Map data Map data | Actin 1 from T. gondii in filaments with MgADP and jasplakinolide; Density modified map from helical reconstruction | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Actin / cytoskeleton / toxoplasmosis / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / cytoskeleton / hydrolase activity / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Hvorecny KL / Sladewski TE / Heaslip AT / Kollman JM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Toxoplasma gondii actin filaments are tuned for rapid disassembly and turnover. Authors: Kelli L Hvorecny / Thomas E Sladewski / Enrique M De La Cruz / Justin M Kollman / Aoife T Heaslip /  Abstract: The cytoskeletal protein actin plays a critical role in the pathogenicity of the intracellular parasite, Toxoplasma gondii, mediating invasion and egress, cargo transport, and organelle inheritance. ...The cytoskeletal protein actin plays a critical role in the pathogenicity of the intracellular parasite, Toxoplasma gondii, mediating invasion and egress, cargo transport, and organelle inheritance. Advances in live cell imaging have revealed extensive filamentous actin networks in the Apicomplexan parasite, but there are conflicting data regarding the biochemical and biophysical properties of Toxoplasma actin. Here, we imaged the in vitro assembly of individual Toxoplasma actin filaments in real time, showing that native, unstabilized filaments grow tens of microns in length. Unlike skeletal muscle actin, Toxoplasma filaments intrinsically undergo rapid treadmilling due to a high critical concentration, fast monomer dissociation, and rapid nucleotide exchange. Cryo-EM structures of jasplakinolide-stabilized and native (i.e. unstabilized) filaments show an architecture like skeletal actin, with differences in assembly contacts in the D-loop that explain the dynamic nature of the filament, likely a conserved feature of Apicomplexan actin. This work demonstrates that evolutionary changes at assembly interfaces can tune the dynamic properties of actin filaments without disrupting their conserved structure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41584.map.gz emd_41584.map.gz | 23.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41584-v30.xml emd-41584-v30.xml emd-41584.xml emd-41584.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41584.png emd_41584.png | 107.3 KB | ||
| Filedesc metadata |  emd-41584.cif.gz emd-41584.cif.gz | 6.2 KB | ||
| Others |  emd_41584_additional_1.map.gz emd_41584_additional_1.map.gz emd_41584_additional_2.map.gz emd_41584_additional_2.map.gz emd_41584_additional_3.map.gz emd_41584_additional_3.map.gz emd_41584_half_map_1.map.gz emd_41584_half_map_1.map.gz emd_41584_half_map_2.map.gz emd_41584_half_map_2.map.gz | 200.7 MB 9.8 MB 200.7 MB 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41584 http://ftp.pdbj.org/pub/emdb/structures/EMD-41584 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41584 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41584 | HTTPS FTP |
-Validation report
| Summary document |  emd_41584_validation.pdf.gz emd_41584_validation.pdf.gz | 753.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41584_full_validation.pdf.gz emd_41584_full_validation.pdf.gz | 753 KB | Display | |
| Data in XML |  emd_41584_validation.xml.gz emd_41584_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_41584_validation.cif.gz emd_41584_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41584 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41584 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41584 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41584 | HTTPS FTP |
-Related structure data
| Related structure data |  8trnMC  8trmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41584.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41584.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin 1 from T. gondii in filaments with MgADP and jasplakinolide; Density modified map from helical reconstruction | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Three protomers of Actin 1 from T. gondii...
| File | emd_41584_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Three protomers of Actin 1 from T. gondii in filaments with MgADP and jasplakinolide; Half map from local refinement | ||||||||||||
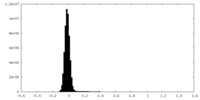
| Projections & Slices |
| ||||||||||||
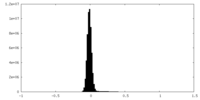
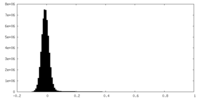
| Density Histograms |
-Additional map: Three protomers of Actin 1 from T. gondii...
| File | emd_41584_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Three protomers of Actin 1 from T. gondii in filaments with MgADP and jasplakinolide; Density modified map from local refinement | ||||||||||||
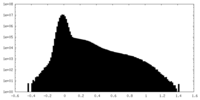
| Projections & Slices |
| ||||||||||||
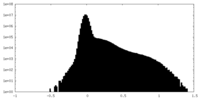
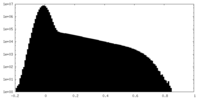
| Density Histograms |
-Additional map: Three protomers of Actin 1 from T. gondii...
| File | emd_41584_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Three protomers of Actin 1 from T. gondii in filaments with MgADP and jasplakinolide; Half map from local refinement | ||||||||||||
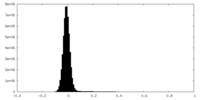
| Projections & Slices |
| ||||||||||||
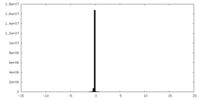
| Density Histograms |
-Half map: Actin 1 from T. gondii in filaments with...
| File | emd_41584_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin 1 from T. gondii in filaments with MgADP and jasplakinolide; Half map from helical reconstruction | ||||||||||||
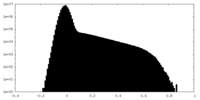
| Projections & Slices |
| ||||||||||||
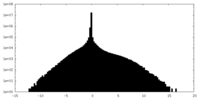
| Density Histograms |
-Half map: Actin 1 from T. gondii in filaments with...
| File | emd_41584_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin 1 from T. gondii in filaments with MgADP and jasplakinolide; Half map from helical reconstruction | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Filament of Actin 1 from T. gondii with MgADP and jasplakinolide
| Entire | Name: Filament of Actin 1 from T. gondii with MgADP and jasplakinolide |
|---|---|
| Components |
|
-Supramolecule #1: Filament of Actin 1 from T. gondii with MgADP and jasplakinolide
| Supramolecule | Name: Filament of Actin 1 from T. gondii with MgADP and jasplakinolide type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Actin protomers from T. gondii assemble into a two-start helix, stabilized by jasplakinolide |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin
| Macromolecule | Name: Actin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.956711 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADEEVQALV VDNGSGNVKA GVAGDDAPRA VFPSIVGKPK NPGIMVGMEE KDCYVGDEAQ SKRGILTLKY PIEHGIVTNW DDMEKIWHH TFYNELRVAP EEHPVLLTEA PLNPKANRER MTQIMFETFN VPAMYVAIQA VLSLYSSGRT TGIVLDSGDG V SHTVPIYE ...String: MADEEVQALV VDNGSGNVKA GVAGDDAPRA VFPSIVGKPK NPGIMVGMEE KDCYVGDEAQ SKRGILTLKY PIEHGIVTNW DDMEKIWHH TFYNELRVAP EEHPVLLTEA PLNPKANRER MTQIMFETFN VPAMYVAIQA VLSLYSSGRT TGIVLDSGDG V SHTVPIYE GYALPHAIMR LDLAGRDLTE YMMKILHERG YGFTTSAEKE IVRDIKEKLC YIALDFDEEM KAAEDSSDIE KS YELPDGN IITVGNERFR CPEALFQPSF LGKEAAGVHR TTFDSIMKCD VDIRKDLYGN VVLSGGTTMY EGIGERLTKE LTS LAPSTM KIKVVAPPER KYSVWIGGSI LSSLSTFQQM WITKEEYDES GPSIVHRKCF UniProtKB: Actin |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 3 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: Jasplakinolide
| Macromolecule | Name: Jasplakinolide / type: ligand / ID: 4 / Number of copies: 3 / Formula: 9UE |
|---|---|
| Molecular weight | Theoretical: 709.67 Da |
| Chemical component information |  ChemComp-9UE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 5.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 45000 |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 28.1 Å Applied symmetry - Helical parameters - Δ&Phi: -167 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PHENIX (ver. 1.20) / Number images used: 2682453 |
|---|---|
| Startup model | Type of model: NONE |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 4.2) |
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8trn: |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X