+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | mGluR3 class 1 in the presence of the antagonist LY 341495 | ||||||||||||
 Map data Map data | Full length map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | GPCR / synaptic protein / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationClass C/3 (Metabotropic glutamate/pheromone receptors) / group II metabotropic glutamate receptor activity / G protein-coupled glutamate receptor signaling pathway / astrocyte projection / G alpha (i) signalling events / postsynaptic modulation of chemical synaptic transmission / calcium channel regulator activity / regulation of synaptic transmission, glutamatergic / sensory perception of pain / modulation of chemical synaptic transmission ...Class C/3 (Metabotropic glutamate/pheromone receptors) / group II metabotropic glutamate receptor activity / G protein-coupled glutamate receptor signaling pathway / astrocyte projection / G alpha (i) signalling events / postsynaptic modulation of chemical synaptic transmission / calcium channel regulator activity / regulation of synaptic transmission, glutamatergic / sensory perception of pain / modulation of chemical synaptic transmission / presynaptic membrane / gene expression / scaffold protein binding / postsynaptic membrane / dendritic spine / postsynaptic density / neuron projection / axon / glutamatergic synapse / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
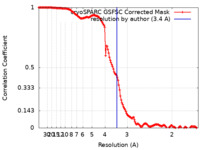
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Strauss A / Levitz J | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis of positive allosteric modulation of metabotropic glutamate receptor activation and internalization. Authors: Alexa Strauss / Alberto J Gonzalez-Hernandez / Joon Lee / Nohely Abreu / Purushotham Selvakumar / Leslie Salas-Estrada / Melanie Kristt / Anisul Arefin / Kevin Huynh / Dagan C Marx / Kristen ...Authors: Alexa Strauss / Alberto J Gonzalez-Hernandez / Joon Lee / Nohely Abreu / Purushotham Selvakumar / Leslie Salas-Estrada / Melanie Kristt / Anisul Arefin / Kevin Huynh / Dagan C Marx / Kristen Gilliland / Bruce J Melancon / Marta Filizola / Joel Meyerson / Joshua Levitz /  Abstract: The metabotropic glutamate receptors (mGluRs) are neuromodulatory family C G protein coupled receptors which assemble as dimers and allosterically couple extracellular ligand binding domains (LBDs) ...The metabotropic glutamate receptors (mGluRs) are neuromodulatory family C G protein coupled receptors which assemble as dimers and allosterically couple extracellular ligand binding domains (LBDs) to transmembrane domains (TMDs) to drive intracellular signaling. Pharmacologically, mGluRs can be targeted at the LBDs by glutamate and synthetic orthosteric compounds or at the TMDs by allosteric modulators. Despite the potential of allosteric compounds as therapeutics, an understanding of the functional and structural basis of their effects is limited. Here we use multiple approaches to dissect the functional and structural effects of orthosteric versus allosteric ligands. We find, using electrophysiological and live cell imaging assays, that both agonists and positive allosteric modulators (PAMs) can drive activation and internalization of group II and III mGluRs. The effects of PAMs are pleiotropic, boosting the maximal response to orthosteric agonists and serving independently as internalization-biased agonists across mGluR subtypes. Motivated by this and intersubunit FRET analyses, we determine cryo-electron microscopy structures of mGluR3 in the presence of either an agonist or antagonist alone or in combination with a PAM. These structures reveal PAM-driven re-shaping of intra- and inter-subunit conformations and provide evidence for a rolling TMD dimer interface activation pathway that controls G protein and beta-arrestin coupling. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41578.map.gz emd_41578.map.gz | 137.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41578-v30.xml emd-41578-v30.xml emd-41578.xml emd-41578.xml | 29.7 KB 29.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41578_fsc.xml emd_41578_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_41578.png emd_41578.png | 45.9 KB | ||
| Filedesc metadata |  emd-41578.cif.gz emd-41578.cif.gz | 6.9 KB | ||
| Others |  emd_41578_additional_1.map.gz emd_41578_additional_1.map.gz emd_41578_additional_2.map.gz emd_41578_additional_2.map.gz emd_41578_additional_3.map.gz emd_41578_additional_3.map.gz emd_41578_additional_4.map.gz emd_41578_additional_4.map.gz emd_41578_additional_5.map.gz emd_41578_additional_5.map.gz emd_41578_additional_6.map.gz emd_41578_additional_6.map.gz emd_41578_half_map_1.map.gz emd_41578_half_map_1.map.gz emd_41578_half_map_2.map.gz emd_41578_half_map_2.map.gz | 253.2 MB 253.2 MB 208.7 MB 207.9 MB 132.7 MB 128 MB 253.4 MB 253.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41578 http://ftp.pdbj.org/pub/emdb/structures/EMD-41578 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41578 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41578 | HTTPS FTP |
-Validation report
| Summary document |  emd_41578_validation.pdf.gz emd_41578_validation.pdf.gz | 941.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41578_full_validation.pdf.gz emd_41578_full_validation.pdf.gz | 941.2 KB | Display | |
| Data in XML |  emd_41578_validation.xml.gz emd_41578_validation.xml.gz | 23 KB | Display | |
| Data in CIF |  emd_41578_validation.cif.gz emd_41578_validation.cif.gz | 30 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41578 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41578 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41578 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41578 | HTTPS FTP |
-Related structure data
| Related structure data |  8trdMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41578.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41578.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full length map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Half map A for locally refined map of the extracellular domain
| File | emd_41578_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A for locally refined map of the extracellular domain | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half map B for locally refined map of the extracellular domain
| File | emd_41578_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B for locally refined map of the extracellular domain | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half map A of the transmembrane domain following particle subtraction
| File | emd_41578_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of the transmembrane domain following particle subtraction | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half map B of the transmembrane domain following particle subtraction
| File | emd_41578_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of the transmembrane domain following particle subtraction | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Locally refined map of the extracellular domain
| File | emd_41578_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally refined map of the extracellular domain | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map of the transmembrane domain following particle subtraction
| File | emd_41578_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the transmembrane domain following particle subtraction | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B of full length map
| File | emd_41578_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of full length map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A of full length map
| File | emd_41578_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of full length map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Metabotropic Glutamate Receptor 3 dimer
| Entire | Name: Metabotropic Glutamate Receptor 3 dimer |
|---|---|
| Components |
|
-Supramolecule #1: Metabotropic Glutamate Receptor 3 dimer
| Supramolecule | Name: Metabotropic Glutamate Receptor 3 dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 260 KDa |
-Macromolecule #1: Metabotropic glutamate receptor 3
| Macromolecule | Name: Metabotropic glutamate receptor 3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 103.161555 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKMLTRLQIL MLALFSKGFL LSLGDHNFMR REIKIEGDLV LGGLFPINEK GTGTEECGRI NEDRGIQRLE AMLFAIDEIN KDNYLLPGV KLGVHILDTC SRDTYALEQS LEFVRASLTK VDEAEYMCPD GSYAIQENIP LLIAGVIGGS YSSVSIQVAN L LRLFQIPQ ...String: MKMLTRLQIL MLALFSKGFL LSLGDHNFMR REIKIEGDLV LGGLFPINEK GTGTEECGRI NEDRGIQRLE AMLFAIDEIN KDNYLLPGV KLGVHILDTC SRDTYALEQS LEFVRASLTK VDEAEYMCPD GSYAIQENIP LLIAGVIGGS YSSVSIQVAN L LRLFQIPQ ISYASTSAKL SDKSRYDYFA RTVPPDFYQA KAMAEILRFF NWTYVSTVAS EGDYGETGIE AFEQEARLRN IC IATAEKV GRSNIRKSYD SVIRELLQKP NARVVVLFMR SDDSRELIAA ANRVNASFTW VASDGWGAQE SIVKGSEHVA YGA ITLELA SHPVRQFDRY FQSLNPYNNH RNPWFRDFWE QKFQCSLQNK RNHRQVCDKH LAIDSSNYEQ ESKIMFVVNA VYAM AHALH KMQRTLCPNT TKLCDAMKIL DGKKLYKEYL LKINFTAPFN PNKGADSIVK FDTFGDGMGR YNVFNLQQTG GKYSY LKVG HWAETLSLDV DSIHWSRNSV PTSQCSDPCA PNEMKNMQPG DVCCWICIPC EPYEYLVDEF TCMDCGPGQW PTADLS GCY NLPEDYIKWE DAWAIGPVTI ACLGFLCTCI VITVFIKHNN TPLVKASGRE LCYILLFGVS LSYCMTFFFI AKPSPVI CA LRRLGLGTSF AICYSALLTK TNCIARIFDG VKNGAQRPKF ISPSSQVFIC LGLILVQIVM VSVWLILETP GTRRYTLP E KRETVILKCN VKDSSMLISL TYDVVLVILC TVYAFKTRKC PENFNEAKFI GFTMYTTCII WLAFLPIFYV TSSDYRVQT TTMCISVSLS GFVVLGCLFA PKVHIVLFQP QKNVVTHRLH LNRFSVSGTA TTYSQSSAST YVPTVCNGRE VLDSTTSSLA GLVPRGSAA AKSAWSHPQF EKGGGSGGGS GGGSWSHPQF EK UniProtKB: Metabotropic glutamate receptor 3 |
-Macromolecule #2: 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine
| Macromolecule | Name: 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine type: ligand / ID: 2 / Number of copies: 2 / Formula: Z99 |
|---|---|
| Molecular weight | Theoretical: 353.369 Da |
| Chemical component information | 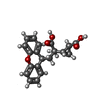 ChemComp-Z99: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8trd: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)