[English] 日本語
 Yorodumi
Yorodumi- EMDB-41301: Zophobas morio black wasting virus strain OR-molitor virion structure -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Zophobas morio black wasting virus strain OR-molitor virion structure | |||||||||
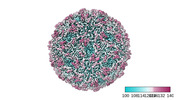
 Map data Map data | Main cyoEM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Capsid / Virion / Parvovirus / Densovirus / Invertebrate / Insect / Pathogen / ssDNA / VIRUS / VIRUS-DNA complex | |||||||||
| Biological species |  Tenebrio molitor (yellow mealworm) / Tenebrio molitor (yellow mealworm) /  Zophobas morio densovirus Zophobas morio densovirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Penzes JJ / Kaelber JT | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Cryo-EM-based discovery of a pathogenic parvovirus causing epidemic mortality by black wasting disease in farmed beetles. Authors: Judit J Penzes / Martin Holm / Samantha A Yost / Jason T Kaelber /  Abstract: We use cryoelectron microscopy (cryo-EM) as a sequence- and culture-independent diagnostic tool to identify the etiological agent of an agricultural pandemic. For the past 4 years, American insect- ...We use cryoelectron microscopy (cryo-EM) as a sequence- and culture-independent diagnostic tool to identify the etiological agent of an agricultural pandemic. For the past 4 years, American insect-rearing facilities have experienced a distinctive larval pathology and colony collapse of farmed Zophobas morio (superworm). By means of cryo-EM, we discovered the causative agent: a densovirus that we named Zophobas morio black wasting virus (ZmBWV). We confirmed the etiology of disease by fulfilling Koch's postulates and characterizing strains from across the United States. ZmBWV is a member of the family Parvoviridae with a 5,542 nt genome, and we describe intersubunit interactions explaining its expanded internal volume relative to human parvoviruses. Cryo-EM structures at resolutions up to 2.1 Å revealed single-strand DNA (ssDNA) ordering at the capsid inner surface pinned by base-binding pockets in the capsid inner surface. Also, we demonstrated the prophylactic potential of non-pathogenic strains to provide cross-protection in vivo. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41301.map.gz emd_41301.map.gz | 1.8 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41301-v30.xml emd-41301-v30.xml emd-41301.xml emd-41301.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41301.png emd_41301.png | 78.6 KB | ||
| Filedesc metadata |  emd-41301.cif.gz emd-41301.cif.gz | 6.6 KB | ||
| Others |  emd_41301_half_map_1.map.gz emd_41301_half_map_1.map.gz emd_41301_half_map_2.map.gz emd_41301_half_map_2.map.gz | 1.6 GB 1.6 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41301 http://ftp.pdbj.org/pub/emdb/structures/EMD-41301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41301 | HTTPS FTP |
-Related structure data
| Related structure data |  8tjeMC  8t9cC  8t9eC  8t9xC  8ta7C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41301.map.gz / Format: CCP4 / Size: 2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41301.map.gz / Format: CCP4 / Size: 2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main cyoEM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.63 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41301_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_41301_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Zophobas morio densovirus
| Entire | Name:  Zophobas morio densovirus Zophobas morio densovirus |
|---|---|
| Components |
|
-Supramolecule #1: Zophobas morio densovirus
| Supramolecule | Name: Zophobas morio densovirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Purified from asymptomatic T. molitor larvae / NCBI-ID: 2750924 / Sci species name: Zophobas morio densovirus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Zophobas morio (beetle) Zophobas morio (beetle) |
| Virus shell | Shell ID: 1 / Diameter: 28.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tenebrio molitor (yellow mealworm) Tenebrio molitor (yellow mealworm) |
| Molecular weight | Theoretical: 48.114625 KDa |
| Sequence | String: ATAILRPIGL HVEKFQQTYR KKWRFLTSAN ANVILAEAAS GERPARWALT TGMASIPWEY LFFYMSPAEY NRMKNYPGTF AKSASVRIR TWNTRVAFQT GDTQTANATL NQNKFLQVAK GIRSIPFICS TNRKYTYSDT EPMQPTGFAT LTSYEYRDGL K TAMYGYDN ...String: ATAILRPIGL HVEKFQQTYR KKWRFLTSAN ANVILAEAAS GERPARWALT TGMASIPWEY LFFYMSPAEY NRMKNYPGTF AKSASVRIR TWNTRVAFQT GDTQTANATL NQNKFLQVAK GIRSIPFICS TNRKYTYSDT EPMQPTGFAT LTSYEYRDGL K TAMYGYDN DNKDFAKKPP ADATGAEIYL QDYLTIYTND ARATTGTKIL AGFPPYKNFI EEFDASACIN TDVVAMDYDF SY APLVPQF APVPNNLITQ NYNGSYPAGT KNEVTAAKTT DSSQATAPTQ VRNAPRKYIQ GPHADTTFFD EEQNYLRVPI EQG GIFEEV NVETVHDTQM PSINVGIRAV PKLTTIDETT QANSWLDAQG YFEVDCVLTT ESVDPYTYIK GGCYSANTKS QLQY FASDG RPIAKVYDNP NVYGRMQMIK TVKP |
-Macromolecule #2: DNA (5'-D(P*CP*G)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*G)-3') / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Tenebrio molitor (yellow mealworm) Tenebrio molitor (yellow mealworm) |
| Molecular weight | Theoretical: 573.43 Da |
| Sequence | String: (DC)(DG) |
-Macromolecule #3: DNA (5'-D(P*CP*AP*GP*GP*CP*CP*AP*AP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*AP*GP*GP*CP*CP*AP*AP*A)-3') / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Tenebrio molitor (yellow mealworm) Tenebrio molitor (yellow mealworm) |
| Molecular weight | Theoretical: 2.733827 KDa |
| Sequence | String: (DC)(DA)(DG)(DG)(DC)(DC)(DA)(DA)(DA) |
-Macromolecule #4: DNA (5'-D(P*TP*CP*GP*AP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*CP*GP*AP*A)-3') / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Tenebrio molitor (yellow mealworm) Tenebrio molitor (yellow mealworm) |
| Molecular weight | Theoretical: 1.504037 KDa |
| Sequence | String: (DT)(DC)(DG)(DA)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Formula: PBS / Component - Name: Phosphate-buffered saline |
| Grid | Support film - Material: GOLD / Support film - topology: HOLEY / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | Purified virus from homogenized T. molitor larval tissue |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 3.0 sec. / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8tje: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)