+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the DHA bound FFA1-Gq complex | |||||||||
 Map data Map data | DHA-FFA1-miniGq | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / agonist / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbioactive lipid receptor activity / Free fatty acid receptors / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Synthesis, secretion, and inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) / response to fatty acid / positive regulation of calcium ion transport / insulin secretion / negative regulation of interleukin-1 beta production / ligand-gated ion channel signaling pathway / positive regulation of insulin secretion ...bioactive lipid receptor activity / Free fatty acid receptors / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Synthesis, secretion, and inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) / response to fatty acid / positive regulation of calcium ion transport / insulin secretion / negative regulation of interleukin-1 beta production / ligand-gated ion channel signaling pathway / positive regulation of insulin secretion / G protein-coupled receptor activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / glucose homeostasis / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / lipid binding / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.39 Å | |||||||||
 Authors Authors | Zhang X / Tikhonova I / Milligan G / Zhang C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Structural basis for the ligand recognition and signaling of free fatty acid receptors. Authors: Xuan Zhang / Abdul-Akim Guseinov / Laura Jenkins / Kunpeng Li / Irina G Tikhonova / Graeme Milligan / Cheng Zhang /   Abstract: Free fatty acid receptors 1 to 4 (FFA1 to FFA4) are class A G protein-coupled receptors (GPCRs). FFA1 to FFA3 share substantial sequence similarity, whereas FFA4 is unrelated. However, FFA1 and FFA4 ...Free fatty acid receptors 1 to 4 (FFA1 to FFA4) are class A G protein-coupled receptors (GPCRs). FFA1 to FFA3 share substantial sequence similarity, whereas FFA4 is unrelated. However, FFA1 and FFA4 are activated by long-chain fatty acids, while FFA2 and FFA3 respond to short-chain fatty acids generated by intestinal microbiota. FFA1, FFA2, and FFA4 are potential drug targets for metabolic and inflammatory conditions. Here, we determined the active structures of FFA1 and FFA4 bound to docosahexaenoic acid, FFA4 bound to the synthetic agonist TUG-891, and butyrate-bound FFA2, each complexed with an engineered heterotrimeric G protein (miniG), by cryo-electron microscopy. Together with computational simulations and mutagenesis studies, we elucidated the similarities and differences in the binding modes of fatty acid ligands to their respective GPCRs. Our findings unveiled distinct mechanisms of receptor activation and G protein coupling. We anticipate that these outcomes will facilitate structure-based drug development and underpin future research on this group of GPCRs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41013.map.gz emd_41013.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41013-v30.xml emd-41013-v30.xml emd-41013.xml emd-41013.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41013.png emd_41013.png | 35.9 KB | ||
| Filedesc metadata |  emd-41013.cif.gz emd-41013.cif.gz | 6.4 KB | ||
| Others |  emd_41013_half_map_1.map.gz emd_41013_half_map_1.map.gz emd_41013_half_map_2.map.gz emd_41013_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41013 http://ftp.pdbj.org/pub/emdb/structures/EMD-41013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41013 | HTTPS FTP |
-Related structure data
| Related structure data |  8t3vMC  8t3oC  8t3qC  8t3sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41013.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41013.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DHA-FFA1-miniGq | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
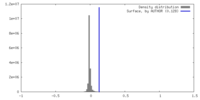
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: DHA-FFA1-miniGq-half B
| File | emd_41013_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DHA-FFA1-miniGq-half_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
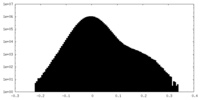
| Density Histograms |
-Half map: DHA-FFA1-miniGq-half A
| File | emd_41013_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DHA-FFA1-miniGq-half_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DHA-FFA1-miniGq complex
| Entire | Name: DHA-FFA1-miniGq complex |
|---|---|
| Components |
|
-Supramolecule #1: DHA-FFA1-miniGq complex
| Supramolecule | Name: DHA-FFA1-miniGq complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(q) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(q) subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.569172 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VSAEDKAAAE RSKMIDKNLR EDGEKARRTL RLLLLGADNS GKSTIVKQMS GIFETKFQVD KVNFHMFDVG GQRDERRKWI QCFNDVTAI IFVVDSSDYN RLQEALNDFK SIWNNRWLRT ISVILFLNKQ DLLAEKVLAG KSKIEDYFPE FARYTTPEDA T PEPGEDPR ...String: VSAEDKAAAE RSKMIDKNLR EDGEKARRTL RLLLLGADNS GKSTIVKQMS GIFETKFQVD KVNFHMFDVG GQRDERRKWI QCFNDVTAI IFVVDSSDYN RLQEALNDFK SIWNNRWLRT ISVILFLNKQ DLLAEKVLAG KSKIEDYFPE FARYTTPEDA T PEPGEDPR VTRAKYFIRK EFVDISTASG DGRHICYPHF TCAVDTENAR RIFNDCKDII LQMNLREYNL V |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.516941 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW ...String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NAICFFPNGN AF ATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGHDNRVSCL GVT DDGMAV ATGSWDSFLK IWNGSS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.261229 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TASIAQARKL VEQLKMEANI DRIKVSKAAA DLMAYCEAHA KEDPLLTPVP ASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.634797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL LEENLYFQGA SHHHHHHHH |
-Macromolecule #5: Free fatty acid receptor 1
| Macromolecule | Name: Free fatty acid receptor 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.395521 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDLPPQLSFG LYVAAFALGF PLNVLAIRGA TAHARLRLTP SLVYALNLGC SDLLLTVSLP LKAVEALASG AWPLPASLCP VFAVAHFFP LYAGGGFLAA LSAGRYLGAA FPLGYQAFRR PCYSWGVCAA IWALVLCHLG LVFGLEAPGG WLDHSNTSLG I NTPVNGSP ...String: MDLPPQLSFG LYVAAFALGF PLNVLAIRGA TAHARLRLTP SLVYALNLGC SDLLLTVSLP LKAVEALASG AWPLPASLCP VFAVAHFFP LYAGGGFLAA LSAGRYLGAA FPLGYQAFRR PCYSWGVCAA IWALVLCHLG LVFGLEAPGG WLDHSNTSLG I NTPVNGSP VCLEAWDPAS AGPARFSLSL LLFFLPLAIT AFCYVGCLRA LARSGLTHRR KLRAAWVAGG ALLTLLLCVG PY NASNVAS FLYPNLGGSW RKLGLITGAW SVVLNPLVTG YL UniProtKB: Free fatty acid receptor 1 |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #7: DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID
| Macromolecule | Name: DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID / type: ligand / ID: 7 / Number of copies: 1 / Formula: HXA |
|---|---|
| Molecular weight | Theoretical: 328.488 Da |
| Chemical component information | 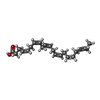 ChemComp-HXA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 61.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.39 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 305318 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)