[English] 日本語
 Yorodumi
Yorodumi- EMDB-40575: Cryo-EM structure of the rat TRPM5 channel in trace calcium, trace-1 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the rat TRPM5 channel in trace calcium, trace-1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / Transient Receptor Potential / TRP / Transient Receptor Potential Melastatin 5 / TRPM5 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcalcium-activated cation channel activity / sodium channel activity / monoatomic ion channel activity / potassium channel activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus / monoatomic ion transmembrane transport / neuronal cell body / dendrite / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
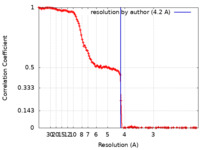
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Karuppan S / Schrag LG / Jara-Oseguera A / Zubcevic L | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel. Authors: Sebastian Karuppan / Lynn Goss Schrag / Caroline M Pastrano / Andrés Jara-Oseguera / Lejla Zubcevic /  Abstract: The transient receptor potential melastatin (TRPM) tetrameric cation channels are involved in a wide range of biological functions, from temperature sensing and taste transduction to regulation of ...The transient receptor potential melastatin (TRPM) tetrameric cation channels are involved in a wide range of biological functions, from temperature sensing and taste transduction to regulation of cardiac function, inflammatory pain, and insulin secretion. The structurally conserved TRPM cytoplasmic domains make up >70 % of the total protein. To investigate the mechanism by which the TRPM cytoplasmic domains contribute to gating, we employed electrophysiology and cryo-EM to study TRPM5-a channel that primarily relies on activation via intracellular Ca. Here, we show that activation of mammalian TRPM5 channels is strongly altered by Ca-dependent desensitization. Structures of rat TRPM5 identify a series of conformational transitions triggered by Ca binding, whereby formation and dissolution of cytoplasmic interprotomer interfaces appear to control activation and desensitization of the channel. This study shows the importance of the cytoplasmic assembly in TRPM5 channel function and sets the stage for future investigations of other members of the TRPM family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40575.map.gz emd_40575.map.gz | 167.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40575-v30.xml emd-40575-v30.xml emd-40575.xml emd-40575.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40575_fsc.xml emd_40575_fsc.xml | 16.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_40575.png emd_40575.png | 72.8 KB | ||
| Filedesc metadata |  emd-40575.cif.gz emd-40575.cif.gz | 6.7 KB | ||
| Others |  emd_40575_additional_1.map.gz emd_40575_additional_1.map.gz emd_40575_additional_2.map.gz emd_40575_additional_2.map.gz emd_40575_additional_3.map.gz emd_40575_additional_3.map.gz emd_40575_additional_4.map.gz emd_40575_additional_4.map.gz emd_40575_half_map_1.map.gz emd_40575_half_map_1.map.gz emd_40575_half_map_2.map.gz emd_40575_half_map_2.map.gz | 167.5 MB 85.7 MB 164.7 MB 164.7 MB 165.2 MB 165.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40575 http://ftp.pdbj.org/pub/emdb/structures/EMD-40575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40575 | HTTPS FTP |
-Related structure data
| Related structure data |  8sl8MC  8sl6C  8slaC  8sleC  8sliC  8slpC  8slqC  8slwC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40575.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40575.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
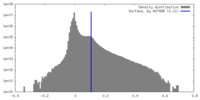
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Focused map for cytosolic domains, sharp
| File | emd_40575_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map for cytosolic domains, sharp | ||||||||||||
| Projections & Slices |
| ||||||||||||
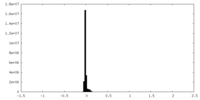
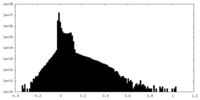
| Density Histograms |
-Additional map: Focused map for cytosolic domains, not sharpened
| File | emd_40575_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map for cytosolic domains, not sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
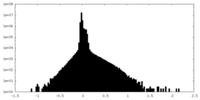
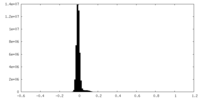
| Density Histograms |
-Additional map: Focused map for cytosolic domains, half map A
| File | emd_40575_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map for cytosolic domains, half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
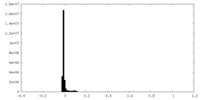
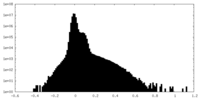
| Density Histograms |
-Additional map: Focused map for cytosolic domains, half map B
| File | emd_40575_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map for cytosolic domains, half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
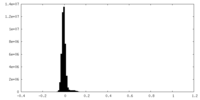
| Density Histograms |
-Half map: half map A
| File | emd_40575_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_40575_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Transient Receptor Potential Melastatin 5 (TRPM5)
| Entire | Name: Transient Receptor Potential Melastatin 5 (TRPM5) |
|---|---|
| Components |
|
-Supramolecule #1: Transient Receptor Potential Melastatin 5 (TRPM5)
| Supramolecule | Name: Transient Receptor Potential Melastatin 5 (TRPM5) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 520 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily M member 5
| Macromolecule | Name: Transient receptor potential cation channel subfamily M member 5 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 132.351719 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKL EMPMAQSSCP GSPPDTGDGW EPVLCKGEVN FGGSGKKRSK FVKVPSNVAP SMLFELLLTE WHLPAPNLVV SLVGEERLF AMKSWLRDVL RKGLVKAAQS TGAWILTSAL HVGLARHVGQ AVRDHSLAST STKVRVVAIG MASLDRILHR Q LLDGVQAQ ...String: MDYKDDDDKL EMPMAQSSCP GSPPDTGDGW EPVLCKGEVN FGGSGKKRSK FVKVPSNVAP SMLFELLLTE WHLPAPNLVV SLVGEERLF AMKSWLRDVL RKGLVKAAQS TGAWILTSAL HVGLARHVGQ AVRDHSLAST STKVRVVAIG MASLDRILHR Q LLDGVQAQ EDTPIHYPAD EGSTQGPLCP LDSNLSHFIL VEPGTLGSGN DGLAELQLSL EKHISQQRTG YGGTSSIQIP VL CLLVNGD PSTLERMSRA VEQAAPWLIL AGSGGIADVL AALVGQPHLL VPQVTEKQFR EKFPSECFSW EAIVHWTELL QNI AAHPHL LTVYDFEQEG SEDLDTVILK ALVKACKSHS RDAQDYLDEL KLAVAWDRVD IAKSEIFNGD VEWKSCDLEE VMTD ALVSN KPDFVRLFVD SGADMAEFLT YGRLQQLYHS VSPKSLLFEL LERKHEEGRL TLAGLGAQQT RELPVGLPAF SLHEV SRVL KDFLHDACRG FYQDGRRMEE RGPPKRPAGQ KWLPDLSRKS EDPWRDLFLW AVLQNRYEMA TYFWAMGREG VAAALA ACK IIKEMSHLEK EAEVARTMRE AKYEQLALDL FSECYSNSED RAFALLVRRN HSWSRTTCLH LATEADAKAF FAHDGVQ AF LTKIWWGDMA TGTPILRLLG AFTCPALIYT NLISFSEDAP QRMDLEDLQE PDSLDMEKSF LCSHGGQLEK LTEAPRAP G DLGPQAAFLL TRWRKFWGAP VTVFLGNVVM YFAFLFLFSY VLLVDFRPPP QGPSGSEVTL YFWVFTLVLE EIRQGFFTN EDTRLVKKFT LYVEDNWNKC DMVAIFLFIV GVTCRMVPSV FEAGRTVLAI DFMVFTLRLI HIFAIHKQLG PKIIIVERMM KDVFFFLFF LSVWLVAYGV TTQALLHPHD GRLEWIFRRV LYRPYLQIFG QIPLDEIDEA RVNCSLHPLL LDSSASCPNL Y ANWLVILL LVTFLLVTNV LLMNLLIAMF SYTFQVVQGN ADMFWKFQRY HLIVEYHGRP ALAPPFILLS HLSLVLKQVF RK EAQHKQQ HLERDLPDPV DQKIITWETV QKENFLSTME KRRRDSEEEV LRKTAHRVDL IAKYIGGLRE QEKRIKCLES QAN YCMLLL SSMTDTLAPG GTYSSSQNCG RRSQPASARD REYLEAGLPH SDT UniProtKB: Transient receptor potential cation channel subfamily M member 5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 296.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)