+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a frog VMAT2 in an apo conformation | |||||||||
 Map data Map data | XlVMAT2 WT. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Vesicular monoamine transporter / SLC18A2 / MFS / fold / TRANSPORT PROTEIN | |||||||||
| Biological species | ||||||||||
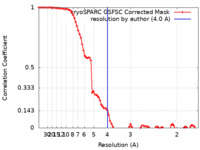
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Lyu Y / Fu C / Ma H / Sun Z / Su Z / Zhou X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Engineering of a mammalian VMAT2 for cryo-EM analysis results in non-canonical protein folding. Authors: Ying Lyu / Chunting Fu / Haiyun Ma / Zhaoming Su / Ziyi Sun / Xiaoming Zhou /  Abstract: Vesicular monoamine transporter 2 (VMAT2) belongs to the major facilitator superfamily (MFS), and mediates cytoplasmic monoamine packaging into presynaptic vesicles. Here, we present two cryo-EM ...Vesicular monoamine transporter 2 (VMAT2) belongs to the major facilitator superfamily (MFS), and mediates cytoplasmic monoamine packaging into presynaptic vesicles. Here, we present two cryo-EM structures of VMAT2, with a frog VMAT2 adopting a canonical MFS fold and an engineered sheep VMAT2 adopting a non-canonical fold. Both VMAT2 proteins mediate uptake of a selective fluorescent VMAT2 substrate into cells. Molecular docking, substrate binding and transport analysis reveal potential substrate binding mechanism in VMAT2. Meanwhile, caution is advised when interpreting engineered membrane protein structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38390.map.gz emd_38390.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38390-v30.xml emd-38390-v30.xml emd-38390.xml emd-38390.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38390_fsc.xml emd_38390_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_38390.png emd_38390.png | 85.7 KB | ||
| Filedesc metadata |  emd-38390.cif.gz emd-38390.cif.gz | 5.7 KB | ||
| Others |  emd_38390_half_map_1.map.gz emd_38390_half_map_1.map.gz emd_38390_half_map_2.map.gz emd_38390_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38390 http://ftp.pdbj.org/pub/emdb/structures/EMD-38390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38390 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38390.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38390.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | XlVMAT2 WT. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map B.
| File | emd_38390_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B. | ||||||||||||
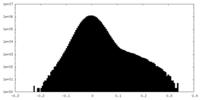
| Projections & Slices |
| ||||||||||||
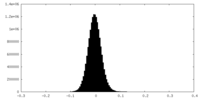
| Density Histograms |
-Half map: Half map A.
| File | emd_38390_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A. | ||||||||||||
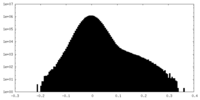
| Projections & Slices |
| ||||||||||||
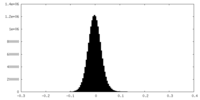
| Density Histograms |
- Sample components
Sample components
-Entire : XlVMAT2 WT
| Entire | Name: XlVMAT2 WT |
|---|---|
| Components |
|
-Supramolecule #1: XlVMAT2 WT
| Supramolecule | Name: XlVMAT2 WT / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #1: XlVMAT2
| Macromolecule | Name: XlVMAT2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 55.624984 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MSMTEFTLLR WLREGRQSRK LILLIVFIAL LLDNMLLTVV VPIIPSYLYS MTHESNNTHD GSIPPAAPSG FHSIFSYYDN TTVITTNFT SSDQLQQSAL TPTTSPSLAT PPASADCPKA DSSLLNENVK VGLLFASKAT VQLLTNPFIG PMTNRIGYQI P MFAGFCIM ...String: MSMTEFTLLR WLREGRQSRK LILLIVFIAL LLDNMLLTVV VPIIPSYLYS MTHESNNTHD GSIPPAAPSG FHSIFSYYDN TTVITTNFT SSDQLQQSAL TPTTSPSLAT PPASADCPKA DSSLLNENVK VGLLFASKAT VQLLTNPFIG PMTNRIGYQI P MFAGFCIM FVSTIMFAFS GTYTLLFIAR SLQGIGSSCS SVAGMGMLAS VYTDDEERGN AMGVALGGLA MGVLVGPPFG SI LYEFVGK TAPFIVLAVL VLFDGALQLF VLQPSRVQPE SQTGTPLLTL IRDPYILIAA GSICFANMAI AMLEPALPIW MME TMCSRK WQLGVAFLPA SISYLLGTNI FGPLAHKMGR WLCAFIGMIM VGISIICVPF ARNIYGLIAP NFGVGFAIGM VDSS MMPIM GYLVDLRHVS VYGSVYAIAD VAFCMGFAFG PSAGGAIAKS IGFPWLMTII GVVDILFAPL CLFLRSPPAR EEKMA ILMD HKCPVKTKMY TQNSGQPYYT GEEEESESDE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8.0 mg/mL |
|---|---|
| Buffer | pH: 6 / Details: 150 mm NaCl, 20 mM MES-Na pH 6.0, 0.4 mM DDM |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 3 s before plunging. |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8xiu: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)