[English] 日本語
 Yorodumi
Yorodumi- EMDB-37910: Structure of the SARS-CoV-2 BA.2.86 spike glycoprotein (closed state) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the SARS-CoV-2 BA.2.86 spike glycoprotein (closed state) | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | spike protein / glycoprotein / VIRUS / VIRAL PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
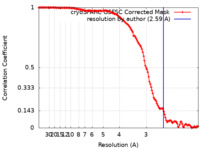
| Method | single particle reconstruction / cryo EM / Resolution: 2.59 Å | |||||||||||||||||||||
 Authors Authors | Yajima H / Anraku Y / Kita S / Kimura K / Maenaka K / Hashiguchi T | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1. Authors: Hisano Yajima / Yuki Anraku / Yu Kaku / Kanako Terakado Kimura / Arnon Plianchaisuk / Kaho Okumura / Yoshiko Nakada-Nakura / Yusuke Atarashi / Takuya Hemmi / Daisuke Kuroda / Yoshimasa ...Authors: Hisano Yajima / Yuki Anraku / Yu Kaku / Kanako Terakado Kimura / Arnon Plianchaisuk / Kaho Okumura / Yoshiko Nakada-Nakura / Yusuke Atarashi / Takuya Hemmi / Daisuke Kuroda / Yoshimasa Takahashi / Shunsuke Kita / Jiei Sasaki / Hiromi Sumita / / Jumpei Ito / Katsumi Maenaka / Kei Sato / Takao Hashiguchi /   Abstract: Since 2019, SARS-CoV-2 has undergone mutations, resulting in pandemic and epidemic waves. The SARS-CoV-2 spike protein, crucial for cellular entry, binds to the ACE2 receptor exclusively when its ...Since 2019, SARS-CoV-2 has undergone mutations, resulting in pandemic and epidemic waves. The SARS-CoV-2 spike protein, crucial for cellular entry, binds to the ACE2 receptor exclusively when its receptor-binding domain (RBD) adopts the up-conformation. However, whether ACE2 also interacts with the RBD in the down-conformation to facilitate the conformational shift to RBD-up remains unclear. Herein, we present the structures of the BA.2.86 and the JN.1 spike proteins bound to ACE2. Notably, we successfully observed the ACE2-bound down-RBD, indicating an intermediate structure before the RBD-up conformation. The wider and mobile angle of RBDs in the up-state provides space for ACE2 to interact with the down-RBD, facilitating the transition to the RBD-up state. The K356T, but not N354-linked glycan, contributes to both of infectivity and neutralizing-antibody evasion in BA.2.86. These structural insights the spike-protein dynamics would help understand the mechanisms underlying SARS-CoV-2 infection and its neutralization. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37910.map.gz emd_37910.map.gz | 108.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37910-v30.xml emd-37910-v30.xml emd-37910.xml emd-37910.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37910_fsc.xml emd_37910_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_37910.png emd_37910.png | 102.3 KB | ||
| Masks |  emd_37910_msk_1.map emd_37910_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37910.cif.gz emd-37910.cif.gz | 7.6 KB | ||
| Others |  emd_37910_additional_1.map.gz emd_37910_additional_1.map.gz emd_37910_half_map_1.map.gz emd_37910_half_map_1.map.gz emd_37910_half_map_2.map.gz emd_37910_half_map_2.map.gz | 108.1 MB 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37910 http://ftp.pdbj.org/pub/emdb/structures/EMD-37910 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37910 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37910 | HTTPS FTP |
-Related structure data
| Related structure data |  8wxlMC  8xuxC  8xuyC  8xuzC  8xv0C  8xv1C  8xvmC  9iu1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37910.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37910.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.005 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37910_msk_1.map emd_37910_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: local refinement closed
| File | emd_37910_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local refinement closed | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37910_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37910_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 BA.2.86 spike glycoprotein
| Entire | Name: SARS-CoV-2 BA.2.86 spike glycoprotein |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 BA.2.86 spike glycoprotein
| Supramolecule | Name: SARS-CoV-2 BA.2.86 spike glycoprotein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 420 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 138.306547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LLMGCVAETG SSQCVMPLFN LITTTQSYTN SFTRGVYYPD KVFRSSVLHL TQDLFLPFFS NVTWFHAISG TNGTKRFDNP VLPFNDGVY FASTEKSNII RGWIFGTTLD SKTQSLLIVN NATNVFIKVC EFQFCNDPFL DVYHKNNKSW MESESGVYSS A NNCTFEYV ...String: LLMGCVAETG SSQCVMPLFN LITTTQSYTN SFTRGVYYPD KVFRSSVLHL TQDLFLPFFS NVTWFHAISG TNGTKRFDNP VLPFNDGVY FASTEKSNII RGWIFGTTLD SKTQSLLIVN NATNVFIKVC EFQFCNDPFL DVYHKNNKSW MESESGVYSS A NNCTFEYV SQPFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPII GRDFPQGFSA LEPLVDLPIG INITRFQTLL AL NRSYLTP GDSSSGWTAG AADYYVGYLQ PRTFLLKYNE NGTITDAVDC ALDPLSETKC TLKSFTVEKG IYQTSNFRVQ PTE SIVRFP NVTNLCPFHE VFNATRFASV YAWNRTRISN CVADYSVLYN FAPFFAFKCY GVSPTKLNDL CFTNVYADSF VIKG NEVSQ IAPGQTGNIA DYNYKLPDDF TGCVIAWNSN KLDSKHSGNY DYWYRLFRKS KLKPFERDIS TEIYQAGNKP CKGKG PNCY FPLQSYGFRP TYGVGHQPYR VVVLSFELLH APATVCGPKK STNLVKNKCV NFNFNGLTGT GVLTKSNKKF LPFQQF GRD IVDTTDAVRD PQTLEILDIT PCSFGGVSVI TPGTNTSNQV AVLYQGVNCT EVSVAIHADQ LTPTWRVYST GSNVFQT RA GCLIGAEYVN NSYECDIPIG AGICASYQTQ TKSRGSAGSV ASQSIIAYTM SLGAENSVAY SNNSIAIPTN FTISVTTE I LPVSMTKTSV DCTMYICGDS TECSNLLLQY GSFCTQLKRA LTGIAVEQDK NTQEVFAQVK QIYKTPPIKY FGGFNFSQI LPDPSKPSKR SPIEDLLFNK VTLADAGFIK QYGDCLGDIA ARDLICAQKF NGLTVLPPLL TDEMIAQYTS ALLAGTITSG WTFGAGPAL QIPFPMQMAY RFNGIGVTQN VLYENQKLIA NQFNSAIGKI QDSLFSTPSA LGKLQDVVNH NAQALNTLVK Q LSSKFGAI SSVLNDILSR LDPPEAEVQI DRLITGRLQS LQTYVTQQLI RAAEIRASAN LAATKMSECV LGQSKRVDFC GK GYHLMSF PQSAPHGVVF LHVTYVPAQE KNFTTAPAIC HDGKAHFPRE GVFVSNGTHW FVTQRNFYEP QIITTDNTFV SGN CDVVIG IVNNTVYDPL QLELDSFKEE LDKYFKNHTS PDVDLGDISG INASVVNIQK EIDRLNEVAK NLNESLIDLQ ELGK YEQYI ASSGYIPEAP RDGQAYVRKD GEWVLLSTFL EGTKHHHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 13 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: blotting time 5 s and blotting force 5.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 3500 / Average exposure time: 1.5 sec. / Average electron dose: 52.45 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)