+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of bsAb3 Fab-Gn-Gc complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN / PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum membrane / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Dabie bandavirus / Dabie bandavirus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
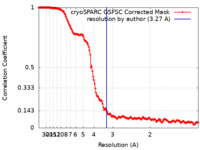
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | |||||||||
 Authors Authors | Wu Y / Sun JQ | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model. Authors: Zhen Chang / Dan Gao / Liying Liao / Junqing Sun / Gen Zhang / Xue Zhang / Feiran Wang / Chunrui Li / Babayemi Olawale Oladejo / Shihua Li / Yan Chai / Yongfei Hu / Xuancheng Lu / Haixia ...Authors: Zhen Chang / Dan Gao / Liying Liao / Junqing Sun / Gen Zhang / Xue Zhang / Feiran Wang / Chunrui Li / Babayemi Olawale Oladejo / Shihua Li / Yan Chai / Yongfei Hu / Xuancheng Lu / Haixia Xiao / Jianxun Qi / Zhihai Chen / Feng Gao / Yan Wu /  Abstract: Severe fever with thrombocytopenia syndrome (SFTS) is an emerging infectious disease with a high fatality rate of up to 30% caused by SFTS virus (SFTSV). However, no specific vaccine or antiviral ...Severe fever with thrombocytopenia syndrome (SFTS) is an emerging infectious disease with a high fatality rate of up to 30% caused by SFTS virus (SFTSV). However, no specific vaccine or antiviral therapy has been approved for clinical use. To develop an effective treatment, we isolated a panel of human monoclonal antibodies (mAbs). SF5 and SF83 are two neutralizing mAbs that recognize two viral glycoproteins (Gn and Gc), respectively. We found that their epitopes are closely located, and we then engineered them as several bispecific antibodies (bsAbs). Neutralization and animal experiments indicated that bsAbs display more potent protective effects than the parental mAbs, and the cryoelectron microscopy structure of a bsAb3 Fab-Gn-Gc complex elucidated the mechanism of protection. In vivo virus passage in the presence of antibodies indicated that two bsAbs resulted in less selective pressure and could efficiently bind to all single parental mAb-escape mutants. Furthermore, epitope analysis of the protective mAbs against SFTSV and RVFV indicated that they are all located on the Gn subdomain I, where may be the hot spots in the phleboviruses. Collectively, these data provide potential therapeutic agents and molecular basis for the rational design of vaccines against SFTSV infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37756.map.gz emd_37756.map.gz | 283.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37756-v30.xml emd-37756-v30.xml emd-37756.xml emd-37756.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37756_fsc.xml emd_37756_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_37756.png emd_37756.png | 19.9 KB | ||
| Filedesc metadata |  emd-37756.cif.gz emd-37756.cif.gz | 7.2 KB | ||
| Others |  emd_37756_half_map_1.map.gz emd_37756_half_map_1.map.gz emd_37756_half_map_2.map.gz emd_37756_half_map_2.map.gz | 301.2 MB 301.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37756 http://ftp.pdbj.org/pub/emdb/structures/EMD-37756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37756 | HTTPS FTP |
-Related structure data
| Related structure data |  8wqwMC  8wsnC  8wspC  8wsuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37756.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37756.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.69 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_37756_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37756_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : the structure of bsAb3_Gn_Gc complex
| Entire | Name: the structure of bsAb3_Gn_Gc complex |
|---|---|
| Components |
|
-Supramolecule #1: the structure of bsAb3_Gn_Gc complex
| Supramolecule | Name: the structure of bsAb3_Gn_Gc complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Gn_Gc
| Supramolecule | Name: Gn_Gc / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1, #4 |
|---|---|
| Source (natural) | Organism:  Dabie bandavirus Dabie bandavirus |
-Supramolecule #3: Fab
| Supramolecule | Name: Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glycoprotein C
| Macromolecule | Name: Glycoprotein C / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dabie bandavirus Dabie bandavirus |
| Molecular weight | Theoretical: 47.204645 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CDEMVHADSK LVSCRQGSGN MKECVTTGRA LLPAVNPGQE ACLHFTAPGS PDSKCLKIKV KRINLKCKKS SSYFVPDARS RCTSVRRCR WAGDCQSGCP PHFTSNSFSD DWAGKMDRAG LGFSGCSDGC GGAACGCFNA APSCIFWRKW VENPHGIIWK V SPCVAWVP ...String: CDEMVHADSK LVSCRQGSGN MKECVTTGRA LLPAVNPGQE ACLHFTAPGS PDSKCLKIKV KRINLKCKKS SSYFVPDARS RCTSVRRCR WAGDCQSGCP PHFTSNSFSD DWAGKMDRAG LGFSGCSDGC GGAACGCFNA APSCIFWRKW VENPHGIIWK V SPCVAWVP SAVIELTMPS GEVRTFHPMS GIPTQVFKGV SVTYLGSDME VSGLTDLCEI EELKSKKLAL APCNQAGMGV VG KVGEIQC SSEESARTIK KDGCIWNADL VGIELRVDDA VCYSKITSVE AVANYSAIPT TIGGLRFERS HDSQGKISGS PLD ITAIRG SFSVNYRGLR LSLSEITATC TGEVTNVSGC YSCMTGAKVS IKLHSSKNST AHVRCKGDET AFSVLEGVHS YTVS LSFDH AVVDEQCQLN CGGHESQVTL KGNLIFLDHH HHHH UniProtKB: Envelopment polyprotein |
-Macromolecule #2: bsAb3 Fab-heavy chain
| Macromolecule | Name: bsAb3 Fab-heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.968289 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVKPGGSLRL SCAASGFTFS NAWMSWVRQA PGKGLEWVGR IKSKTDGGTT DYAAPVKGRF TISRDDSKNT LYLQMNSLK TEDTAVYYCT TDFISAGPWY FDLWGRGTLV TVSSGGGGSG GGGSQVQLVQ SGGGVVKTGR SQRVSCATSG F TFSSYAIH ...String: EVQLVESGGG LVKPGGSLRL SCAASGFTFS NAWMSWVRQA PGKGLEWVGR IKSKTDGGTT DYAAPVKGRF TISRDDSKNT LYLQMNSLK TEDTAVYYCT TDFISAGPWY FDLWGRGTLV TVSSGGGGSG GGGSQVQLVQ SGGGVVKTGR SQRVSCATSG F TFSSYAIH WVRQAPGKGL EWVAVISYDG SNKYYADSVK GRFTISRDSS KNTVYLQMNS LRTEDTAVYY CARSQASWQA FD YWGQGTL VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE PVTVSWNSGA LTSGVHTFPA VLQSSGLYSL SSV VTVPSS SLGTQTYICN VNHKPSNTKV DKKVEPKSCD K |
-Macromolecule #3: bsAb3 Fab-light chain
| Macromolecule | Name: bsAb3 Fab-light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.276727 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SSELTQDPAV SVALGQTVRI TCQGDSLRSY YASWYQQKPG QAPVLVIYGK NNRPSGIPDR FSGSSSGNTA SLTITGAQAE DEADYYCNS RDSSGNHVVF GGGTKLTVLG GGGSGGGGSD IQMTQSPSSL SASVGDRVTI TCRASQGVGT DLAWYQQKPG K APNRLIFA ...String: SSELTQDPAV SVALGQTVRI TCQGDSLRSY YASWYQQKPG QAPVLVIYGK NNRPSGIPDR FSGSSSGNTA SLTITGAQAE DEADYYCNS RDSSGNHVVF GGGTKLTVLG GGGSGGGGSD IQMTQSPSSL SASVGDRVTI TCRASQGVGT DLAWYQQKPG K APNRLIFA ASNLQSGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCLH HNSFPLAFGG GTKVEIKRTV AAPSVFIFPP SD EQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS QESVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLS SPVTKS FNRGECS |
-Macromolecule #4: Glycoprotein N
| Macromolecule | Name: Glycoprotein N / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dabie bandavirus Dabie bandavirus |
| Molecular weight | Theoretical: 35.780461 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DGIQDSGPII CAGPIHSNKS ADIPHLLGYS EKICQIDRLI HVSSWLRNHS QFQGYVGQRG GRSQVSYYPA ENSYSRWSGL LSPCDADWL GMLVVKKAKG SDMIVPGPSY KGKVFFERPT FDGYVGWGCG SGKSRTESGE LCSSDSGTSS GLLPSDRVLW I GDVACQPM ...String: DGIQDSGPII CAGPIHSNKS ADIPHLLGYS EKICQIDRLI HVSSWLRNHS QFQGYVGQRG GRSQVSYYPA ENSYSRWSGL LSPCDADWL GMLVVKKAKG SDMIVPGPSY KGKVFFERPT FDGYVGWGCG SGKSRTESGE LCSSDSGTSS GLLPSDRVLW I GDVACQPM TPIPEETFLE LKSFSQSEFP DICKIDGIVF NQCEGESLPQ PFDVAWMDVG HSHKIIMREH KTKWVQESSS KD FVCYKEG TGPCSESEEK TCKTSGSCRG DMQFCKVAGC EHGEEASEAK CRCSLVHKPG EVVVSYGGMR VRPKCYGFSR MMA TLEVN UniProtKB: Envelopment polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)