[English] 日本語
 Yorodumi
Yorodumi- EMDB-37749: In situ PBS-PSII supercomplex from cyanobacterial Spirulina platensis -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ PBS-PSII supercomplex from cyanobacterial Spirulina platensis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationphycobilisome / plasma membrane-derived thylakoid membrane / photosynthesis Similarity search - Function | |||||||||
| Biological species |  Arthrospira platensis (bacteria) / Arthrospira platensis (bacteria) /  Arthrospira sp. FACHB-439 (bacteria) Arthrospira sp. FACHB-439 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | You X / Zhang X / Xiao YN / Sun S / Sui SF | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: In situ structural determination of cyanobacterial phycobilisome-PSII supercomplex by STAgSPA strategy. Authors: Xing Zhang / Yanan Xiao / Xin You / Shan Sun / Sen-Fang Sui /  Abstract: Photosynthesis converting solar energy to chemical energy is one of the most important chemical reactions on earth. In cyanobacteria, light energy is captured by antenna system phycobilisomes (PBSs) ...Photosynthesis converting solar energy to chemical energy is one of the most important chemical reactions on earth. In cyanobacteria, light energy is captured by antenna system phycobilisomes (PBSs) and transferred to photosynthetic reaction centers of photosystem II (PSII) and photosystem I (PSI). While most of the protein complexes involved in photosynthesis have been characterized by in vitro structural analyses, how these protein complexes function together in vivo is not well understood. Here we implemented STAgSPA, an in situ structural analysis strategy, to solve the native structure of PBS-PSII supercomplex from the cyanobacteria Arthrospira sp. FACHB439 at resolution of ~3.5 Å. The structure reveals coupling details among adjacent PBSs and PSII dimers, and the collaborative energy transfer mechanism mediated by multiple super-PBS in cyanobacteria. Our results provide insights into the diversity of photosynthesis-related systems between prokaryotic cyanobacteria and eukaryotic red algae but are also a methodological demonstration for high-resolution structural analysis in cellular or tissue samples. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37749.map.gz emd_37749.map.gz | 228.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37749-v30.xml emd-37749-v30.xml emd-37749.xml emd-37749.xml | 62.2 KB 62.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37749.png emd_37749.png | 123.6 KB | ||
| Filedesc metadata |  emd-37749.cif.gz emd-37749.cif.gz | 12.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37749 http://ftp.pdbj.org/pub/emdb/structures/EMD-37749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37749 | HTTPS FTP |
-Related structure data
| Related structure data |  8wqlMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37749.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37749.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.632 Å | ||||||||||||||||||||||||||||||||||||
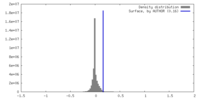
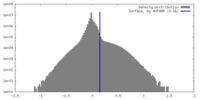
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : In situ PBS-PSII supercomplex
+Supramolecule #1: In situ PBS-PSII supercomplex
+Macromolecule #1: LRC1
+Macromolecule #2: LCM
+Macromolecule #3: Allophycocyanin alpha chain
+Macromolecule #4: Allophycocyanin beta chain
+Macromolecule #5: Phycobilisome 7.8 kDa linker polypeptide, allophycocyanin-associa...
+Macromolecule #6: Photosystem II protein D1
+Macromolecule #7: Photosystem II CP47 reaction center protein
+Macromolecule #8: Photosystem II CP43 reaction center protein
+Macromolecule #9: Photosystem II D2 protein
+Macromolecule #10: Cytochrome b559 subunit alpha
+Macromolecule #11: Cytochrome b559 subunit beta
+Macromolecule #12: Photosystem II reaction center protein H
+Macromolecule #13: Photosystem II reaction center protein I
+Macromolecule #14: Photosystem II reaction center protein J
+Macromolecule #15: Photosystem II reaction center protein K
+Macromolecule #16: Photosystem II reaction center protein L
+Macromolecule #17: Photosystem II reaction center protein M
+Macromolecule #18: Photosystem II extrinsic protein O
+Macromolecule #19: PsbQ protein
+Macromolecule #20: Photosystem II reaction center protein Y
+Macromolecule #21: LPP1
+Macromolecule #22: Photosystem II reaction center protein T
+Macromolecule #23: PsbU
+Macromolecule #24: Photosystem II extrinsic protein V
+Macromolecule #25: Photosystem II reaction center protein X
+Macromolecule #26: Photosystem II reaction center protein Psb30
+Macromolecule #27: Photosystem II reaction center protein Z
+Macromolecule #28: LPP2
+Macromolecule #29: Allophycocyanin beta-18 subunit
+Macromolecule #30: Allophycocyanin-B alpha subunit
+Macromolecule #31: C-phycocyanin alpha subunit
+Macromolecule #32: Phycocyanin beta subunit
+Macromolecule #33: PHYCOCYANOBILIN
+Macromolecule #34: CA-MN4-O5 CLUSTER
+Macromolecule #35: CHLORIDE ION
+Macromolecule #36: CHLOROPHYLL A
+Macromolecule #37: 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,...
+Macromolecule #38: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
+Macromolecule #39: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #40: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #41: BICARBONATE ION
+Macromolecule #42: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #43: BETA-CAROTENE
+Macromolecule #44: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #45: PHEOPHYTIN A
+Macromolecule #46: FE (II) ION
+Macromolecule #47: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #48: CALCIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus min: 6.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)