[English] 日本語
 Yorodumi
Yorodumi- EMDB-36206: Human sodium-dependent vitamin C transporter 1 in an apo occluded... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human sodium-dependent vitamin C transporter 1 in an apo occluded state | |||||||||
 Map data Map data | FSC-weighted, sharpened and masked map by PostProcess | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transporter / Membrane protein / Ascorbic acid / Vitamin C / Sodium / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnucleobase transport / L-ascorbate:sodium symporter activity / L-ascorbic acid transmembrane transporter activity / L-ascorbic acid transmembrane transport / nucleobase transmembrane transporter activity / dehydroascorbic acid transmembrane transporter activity / intracellular organelle / sodium ion transmembrane transporter activity / dehydroascorbic acid transport / Vitamin C (ascorbate) metabolism ...nucleobase transport / L-ascorbate:sodium symporter activity / L-ascorbic acid transmembrane transporter activity / L-ascorbic acid transmembrane transport / nucleobase transmembrane transporter activity / dehydroascorbic acid transmembrane transporter activity / intracellular organelle / sodium ion transmembrane transporter activity / dehydroascorbic acid transport / Vitamin C (ascorbate) metabolism / L-ascorbic acid metabolic process / urate transmembrane transporter activity / sodium ion transport / lung development / basal plasma membrane / brain development / response to toxic substance / apical plasma membrane / extracellular exosome / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | |||||||||
 Authors Authors | Kobayashi TA / Kusakizako T / Nureki O | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Dimeric transport mechanism of human vitamin C transporter SVCT1. Authors: Takaaki A Kobayashi / Hiroto Shimada / Fumiya K Sano / Yuzuru Itoh / Sawako Enoki / Yasushi Okada / Tsukasa Kusakizako / Osamu Nureki /  Abstract: Vitamin C plays important roles as a cofactor in many enzymatic reactions and as an antioxidant against oxidative stress. As some mammals including humans cannot synthesize vitamin C de novo from ...Vitamin C plays important roles as a cofactor in many enzymatic reactions and as an antioxidant against oxidative stress. As some mammals including humans cannot synthesize vitamin C de novo from glucose, its uptake from dietary sources is essential, and is mediated by the sodium-dependent vitamin C transporter 1 (SVCT1). Despite its physiological significance in maintaining vitamin C homeostasis, the structural basis of the substrate transport mechanism remained unclear. Here, we report the cryo-EM structures of human SVCT1 in different states at 2.5-3.5 Å resolutions. The binding manner of vitamin C together with two sodium ions reveals the counter ion-dependent substrate recognition mechanism. Furthermore, comparisons of the inward-open and occluded structures support a transport mechanism combining elevator and distinct rotational motions. Our results demonstrate the molecular mechanism of vitamin C transport with its underlying conformational cycle, potentially leading to future industrial and medical applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36206.map.gz emd_36206.map.gz | 2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36206-v30.xml emd-36206-v30.xml emd-36206.xml emd-36206.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36206_fsc.xml emd_36206_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_36206.png emd_36206.png | 98.8 KB | ||
| Masks |  emd_36206_msk_1.map emd_36206_msk_1.map | 6.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36206.cif.gz emd-36206.cif.gz | 6.7 KB | ||
| Others |  emd_36206_half_map_1.map.gz emd_36206_half_map_1.map.gz emd_36206_half_map_2.map.gz emd_36206_half_map_2.map.gz | 5.5 MB 5.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36206 http://ftp.pdbj.org/pub/emdb/structures/EMD-36206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36206 | HTTPS FTP |
-Related structure data
| Related structure data |  8jf1MC  8jewC  8jezC  8jf0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36206.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36206.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FSC-weighted, sharpened and masked map by PostProcess | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.996 Å | ||||||||||||||||||||||||||||||||||||
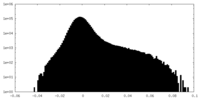
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36206_msk_1.map emd_36206_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
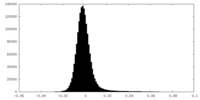
| Density Histograms |
-Half map: #2
| File | emd_36206_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
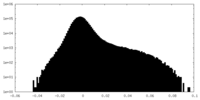
| Density Histograms |
-Half map: #1
| File | emd_36206_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
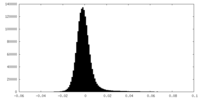
| Density Histograms |
- Sample components
Sample components
-Entire : Human SVCT1 dimer in an apo occluded state
| Entire | Name: Human SVCT1 dimer in an apo occluded state |
|---|---|
| Components |
|
-Supramolecule #1: Human SVCT1 dimer in an apo occluded state
| Supramolecule | Name: Human SVCT1 dimer in an apo occluded state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 23 member 1
| Macromolecule | Name: Solute carrier family 23 member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 64.860707 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRAQEDLEGR TQHETTRDPS TPLPTEPKFD MLYKIEDVPP WYLCILLGFQ HYLTCFSGTI AVPFLLAEAL CVGHDQHMVS QLIGTIFTC VGITTLIQTT VGIRLPLFQA SAFAFLVPAK AILALERWKC PPEEEIYGNW SLPLNTSHIW HPRIREVQGA I MVSSVVEV ...String: MRAQEDLEGR TQHETTRDPS TPLPTEPKFD MLYKIEDVPP WYLCILLGFQ HYLTCFSGTI AVPFLLAEAL CVGHDQHMVS QLIGTIFTC VGITTLIQTT VGIRLPLFQA SAFAFLVPAK AILALERWKC PPEEEIYGNW SLPLNTSHIW HPRIREVQGA I MVSSVVEV VIGLLGLPGA LLNYIGPLTV TPTVSLIGLS VFQAAGDRAG SHWGISACSI LLIILFSQYL RNLTFLLPVY RW GKGLTLL RIQIFKMFPI MLAIMTVWLL CYVLTLTDVL PTDPKAYGFQ ARTDARGDIM AIAPWIRIPY PCQWGLPTVT AAA VLGMFS ATLAGIIESI GDYYACARLA GAPPPPVHAI NRGIFTEGIC CIIAGLLGTG NGSTSSSPNI GVLGITKVGS RRVV QYGAA IMLVLGTIGK FTALFASLPD PILGGMFCTL FGMITAVGLS NLQFVDMNSS RNLFVLGFSM FFGLTLPNYL ESNPG AINT GILEVDQILI VLLTTEMFVG GCLAFILDNT VPGSPEERGL IQWKAGAHAN SDMSSSLKSY DFPIGMGIVK RITFLK YIP ICPVFKGFSS SSKDQIAIPE DTPENTETAS VCTKV UniProtKB: Solute carrier family 23 member 1 |
-Macromolecule #2: Lauryl Maltose Neopentyl Glycol
| Macromolecule | Name: Lauryl Maltose Neopentyl Glycol / type: ligand / ID: 2 / Number of copies: 3 / Formula: AV0 |
|---|---|
| Molecular weight | Theoretical: 1.005188 KDa |
| Chemical component information |  ChemComp-AV0: |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #4: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 4 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 6 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 25 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 5907 / Average electron dose: 50.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)