+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
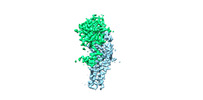
| Title | Cryo-EM structure of yeast Erf2/Erf4 complex | |||||||||
 Map data Map data | the yeast refined map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | palmitoyltransferase / Saccaromyces cerevisiae / palmitoylation / RAS2 / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationendoplasmic reticulum palmitoyltransferase complex / protein palmitoylation / protein S-acyltransferase / protein-cysteine S-palmitoyltransferase activity / palmitoyltransferase activity / perinuclear endoplasmic reticulum / cortical endoplasmic reticulum / protein targeting to membrane / Neutrophil degranulation / endoplasmic reticulum membrane ...endoplasmic reticulum palmitoyltransferase complex / protein palmitoylation / protein S-acyltransferase / protein-cysteine S-palmitoyltransferase activity / palmitoyltransferase activity / perinuclear endoplasmic reticulum / cortical endoplasmic reticulum / protein targeting to membrane / Neutrophil degranulation / endoplasmic reticulum membrane / endoplasmic reticulum / Golgi apparatus Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Wu J / Hu Q / Zhang Y / Yang A / Liu S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation. Authors: Anlan Yang / Shengjie Liu / Yuqi Zhang / Jia Chen / Yujing Fan / Fengxiang Wang / Yilong Zou / Shan Feng / Jianping Wu / Qi Hu /  Abstract: Palmitoylation of cysteine residues at the C-terminal hypervariable regions in human HRAS and NRAS, which is necessary for RAS signaling, is catalyzed by the acyltransferase DHHC9 in complex with its ...Palmitoylation of cysteine residues at the C-terminal hypervariable regions in human HRAS and NRAS, which is necessary for RAS signaling, is catalyzed by the acyltransferase DHHC9 in complex with its accessory protein GCP16. The molecular basis for the acyltransferase activity and the regulation of DHHC9 by GCP16 is not clear. Here we report the cryo-electron microscopy structures of the human DHHC9-GCP16 complex and its yeast counterpart-the Erf2-Erf4 complex, demonstrating that GCP16 and Erf4 are not directly involved in the catalytic process but stabilize the architecture of DHHC9 and Erf2, respectively. We found that a phospholipid binding to an arginine-rich region of DHHC9 and palmitoylation on three residues (C24, C25 and C288) were essential for the catalytic activity of the DHHC9-GCP16 complex. Moreover, we showed that GCP16 also formed complexes with DHHC14 and DHHC18 to catalyze RAS palmitoylation. These findings provide insights into the regulatory mechanism of RAS palmitoyltransferases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34717.map.gz emd_34717.map.gz | 25.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34717-v30.xml emd-34717-v30.xml emd-34717.xml emd-34717.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34717.png emd_34717.png | 21.7 KB | ||
| Filedesc metadata |  emd-34717.cif.gz emd-34717.cif.gz | 6.4 KB | ||
| Others |  emd_34717_half_map_1.map.gz emd_34717_half_map_1.map.gz emd_34717_half_map_2.map.gz emd_34717_half_map_2.map.gz | 25.1 MB 25.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34717 http://ftp.pdbj.org/pub/emdb/structures/EMD-34717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34717 | HTTPS FTP |
-Related structure data
| Related structure data |  8hfcMC  8hf3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34717.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34717.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | the yeast refined map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B
| File | emd_34717_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_34717_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Erf2/Erf4 complex
| Entire | Name: Erf2/Erf4 complex |
|---|---|
| Components |
|
-Supramolecule #1: Erf2/Erf4 complex
| Supramolecule | Name: Erf2/Erf4 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Palmitoyltransferase ERF2
| Macromolecule | Name: Palmitoyltransferase ERF2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: protein S-acyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.811672 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHHH HHHHGSDEVD AHMALVSRRS TRSESTSITK EEHTGEGSLT KLFFRWLVTL EGDQDINDGK GYISLPNVSN YIFFLGGRF RTVKGAKPLW LGVLLAIVCP MVLFSIFEAH KLWHTQNGYK VLVIFFYYFW VITLASFIRT ATSDPGVLPR N IHLSQLRN ...String: MGHHHHHHHH HHHHGSDEVD AHMALVSRRS TRSESTSITK EEHTGEGSLT KLFFRWLVTL EGDQDINDGK GYISLPNVSN YIFFLGGRF RTVKGAKPLW LGVLLAIVCP MVLFSIFEAH KLWHTQNGYK VLVIFFYYFW VITLASFIRT ATSDPGVLPR N IHLSQLRN NYQIPQEYYN LITLPTHSSI SKDITIKYCP SCRIWRPPRS SHCSTCNVCV MVHDHHCIWV NNCIGKRNYR FF LIFLLGA ILSSVILLTN CAIHIARESG GPRDCPVAIL LLCYAGLTLW YPAILFTYHI FMAGNQQTTR EFLKGIGSKK NPV FHRVVK EENIYNKGSF LKNMGHLMLE PRGPSFVSAR KPHEAGDWRF MDLSPAHSFE KIQKI UniProtKB: Palmitoyltransferase ERF2 |
-Macromolecule #2: Ras modification protein ERF4
| Macromolecule | Name: Ras modification protein ERF4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.652705 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKG SDYKDDDDKG SDEVDAHMCD SHQKEEDNAN TSERALFFNY HEFSYSFYED LGSEDAKPTE HDEDHKLCIT HFPNVYAAR GSAEFQVTRV VRVPRRFDES RSSLETPQFS TQLPGSEPAA IVGDDGTSFV RCGRYDIGDH VFGCSSVSPL S EYLSAAEL ...String: MDYKDDDDKG SDYKDDDDKG SDEVDAHMCD SHQKEEDNAN TSERALFFNY HEFSYSFYED LGSEDAKPTE HDEDHKLCIT HFPNVYAAR GSAEFQVTRV VRVPRRFDES RSSLETPQFS TQLPGSEPAA IVGDDGTSFV RCGRYDIGDH VFGCSSVSPL S EYLSAAEL AEVVHRVNGF LLREEGEVFG WRNLSGLLLD MLTGGLWSWV LGPLLSRPVF QESLALEQYV AQLNSPGGLL HE RGVRLVL PRRSGCLSLD FVVPRPK UniProtKB: Ras modification protein ERF4 |
-Macromolecule #3: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 3 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)