[English] 日本語
 Yorodumi
Yorodumi- EMDB-34307: Structure of the intact photosynthetic light-harvesting antenna-r... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Green sulfur bacterium / Reaction center / FMO protein / Energy transfer / PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationthylakoid / bacteriochlorophyll binding / iron-sulfur cluster binding / photosynthesis / electron transfer activity / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Chlorobaculum tepidum TLS (bacteria) Chlorobaculum tepidum TLS (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Chen JH / Zhang X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Integr Plant Biol / Year: 2023 Journal: J Integr Plant Biol / Year: 2023Title: Cryo-electron microscopy structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium. Authors: Jing-Hua Chen / Weiwei Wang / Chen Wang / Tingyun Kuang / Jian-Ren Shen / Xing Zhang /   Abstract: The photosynthetic reaction center complex (RCC) of green sulfur bacteria (GSB) consists of the membrane-imbedded RC core and the peripheric energy transmitting proteins called Fenna-Matthews-Olson ...The photosynthetic reaction center complex (RCC) of green sulfur bacteria (GSB) consists of the membrane-imbedded RC core and the peripheric energy transmitting proteins called Fenna-Matthews-Olson (FMO). Functionally, FMO transfers the absorbed energy from a huge peripheral light-harvesting antenna named chlorosome to the RC core where charge separation occurs. In vivo, one RC was found to bind two FMOs, however, the intact structure of RCC as well as the energy transfer mechanism within RCC remain to be clarified. Here we report a structure of intact RCC which contains a RC core and two FMO trimers from a thermophilic green sulfur bacterium Chlorobaculum tepidum at 2.9 Å resolution by cryo-electron microscopy. The second FMO trimer is attached at the cytoplasmic side asymmetrically relative to the first FMO trimer reported previously. We also observed two new subunits (PscE and PscF) and the N-terminal transmembrane domain of a cytochrome-containing subunit (PscC) in the structure. These two novel subunits possibly function to facilitate the binding of FMOs to RC core and to stabilize the whole complex. A new bacteriochlorophyll (numbered as 816) was identified at the interspace between PscF and PscA-1, causing an asymmetrical energy transfer from the two FMO trimers to RC core. Based on the structure, we propose an energy transfer network within this photosynthetic apparatus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34307.map.gz emd_34307.map.gz | 141.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34307-v30.xml emd-34307-v30.xml emd-34307.xml emd-34307.xml | 37.2 KB 37.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34307.png emd_34307.png | 112.9 KB | ||
| Filedesc metadata |  emd-34307.cif.gz emd-34307.cif.gz | 9.3 KB | ||
| Others |  emd_34307_half_map_1.map.gz emd_34307_half_map_1.map.gz emd_34307_half_map_2.map.gz emd_34307_half_map_2.map.gz | 139.3 MB 139.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34307 http://ftp.pdbj.org/pub/emdb/structures/EMD-34307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34307 | HTTPS FTP |
-Related structure data
| Related structure data |  8gwaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34307.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34307.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34307_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
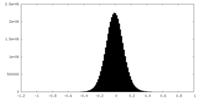
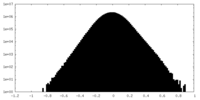
| Density Histograms |
-Half map: #1
| File | emd_34307_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
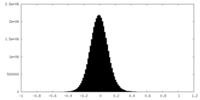
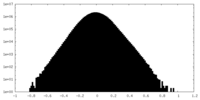
| Density Histograms |
- Sample components
Sample components
+Entire : GsbRC-2FMO complex
+Supramolecule #1: GsbRC-2FMO complex
+Macromolecule #1: Bacteriochlorophyll a protein
+Macromolecule #2: P840 reaction center 17 kDa protein
+Macromolecule #3: Photosystem P840 reaction center iron-sulfur protein
+Macromolecule #4: Photosystem P840 reaction center, large subunit
+Macromolecule #5: unkown protein
+Macromolecule #6: Ric1 protein
+Macromolecule #7: Cytochrome c
+Macromolecule #8: BACTERIOCHLOROPHYLL A
+Macromolecule #9: CARDIOLIPIN
+Macromolecule #10: IRON/SULFUR CLUSTER
+Macromolecule #11: Bacteriochlorophyll A isomer
+Macromolecule #12: Chlorophyll A ester
+Macromolecule #13: [(2R,3S,4S,5R,6R)-6-[(10E,12E,14E)-2,6,10,14,19,23-hexamethyl-25-...
+Macromolecule #14: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #15: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #16: CALCIUM ION
+Macromolecule #17: 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethy...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 90.0 K |
| Specialist optics | Spherical aberration corrector: 2.7mm / Chromatic aberration corrector: 2.7mm / Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 2297 / Average exposure time: 8.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 152200 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)